1G3S
 
 | | CYS102SER DTXR | | Descriptor: | DIPHTHERIA TOXIN REPRESSOR | | Authors: | Pohl, E, Goranson-Siekierke, J, Holmes, R.K, Hol, W.G.J. | | Deposit date: | 2000-10-25 | | Release date: | 2001-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of three diphtheria toxin repressor (DtxR) variants with decreased repressor activity.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1G3W
 
 | | CD-CYS102SER DTXR | | Descriptor: | CADMIUM ION, DIPHTHERIA TOXIN REPRESSOR, SULFATE ION | | Authors: | Pohl, E, Goranson-Siekierke, J, Holmes, R.K, Hol, W.G.J. | | Deposit date: | 2000-10-25 | | Release date: | 2001-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of three diphtheria toxin repressor (DtxR) variants with decreased repressor activity.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
4L0Z
 
 | |
4L0Y
 
 | |
4L18
 
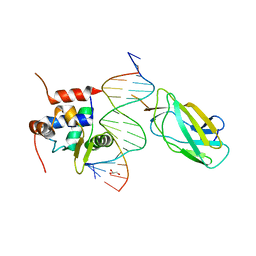 | |
3GBG
 
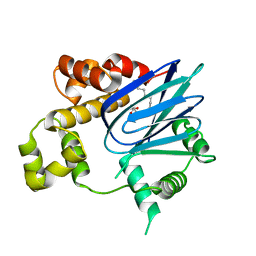 | | Crystal Structure of ToxT from Vibrio Cholerae O395 | | Descriptor: | PALMITOLEIC ACID, TCP pilus virulence regulatory protein | | Authors: | Lowden, M.J, Kull, F.J. | | Deposit date: | 2009-02-19 | | Release date: | 2010-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Vibrio cholerae ToxT reveals a mechanism for fatty acid regulation of virulence genes.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1H9G
 
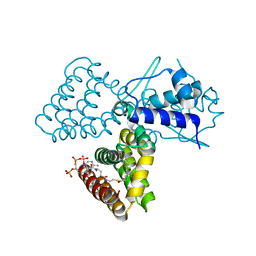 | | FadR, FATTY ACID RESPONSIVE TRANSCRIPTION FACTOR FROM E. COLI, in complex with myristoyl-CoA | | Descriptor: | COENZYME A, FATTY ACID METABOLISM REGULATOR PROTEIN, MYRISTIC ACID | | Authors: | Van Aalten, D.M.F, Dirusso, C.C, Knudsen, J. | | Deposit date: | 2001-03-09 | | Release date: | 2001-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structural Basis of Acyl Coenzyme A-Dependent Regulation of the Transcription Factor Fadr
Embo J., 20, 2001
|
|
1H9T
 
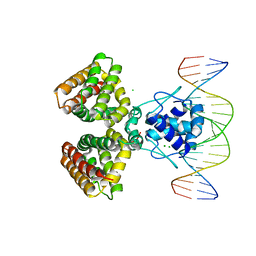 | | FADR, FATTY ACID RESPONSIVE TRANSCRIPTION FACTOR FROM E. COLI IN COMPLEX WITH FADB OPERATOR | | Descriptor: | 5'-D(*CP*AP*TP*CP*TP*GP*GP*TP*AP*CP*GP*AP* CP*CP*AP*GP*AP*TP*C)-3', 5'-D(*GP*AP*TP*CP*TP*GP*GP*TP*CP*GP*TP*AP* CP*CP*AP*GP*AP*TP*G)-3', CHLORIDE ION, ... | | Authors: | Van Aalten, D.M.F, Dirusso, C.C, Knudsen, J. | | Deposit date: | 2001-03-19 | | Release date: | 2001-04-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The Structural Basis of Acyl Coenzyme A-Dependent Regulation of the Transcription Factor Fadr
Embo J., 20, 2001
|
|
8Z50
 
 | | Crystal structure of the ASF1-H3T-H4 complex | | Descriptor: | Histone H3.1t, Histone H4, Histone chaperone ASF1A | | Authors: | Xu, L. | | Deposit date: | 2024-04-18 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into instability of the nucleosome driven by histone variant H3T.
Biochem.Biophys.Res.Commun., 727, 2024
|
|
6LBS
 
 | | Crystal structure of yeast Stn1 | | Descriptor: | KLLA0C11825p | | Authors: | Ge, Y, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2019-11-14 | | Release date: | 2020-07-15 | | Last modified: | 2020-08-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into telomere protection and homeostasis regulation by yeast CST complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6LBT
 
 | | Crystal structure of yeast Cdc13 and Stn1 | | Descriptor: | GLYCEROL, KLLA0C11825p, KLLA0F20922p,KLLA0F20922p | | Authors: | Ge, Y, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2019-11-14 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into telomere protection and homeostasis regulation by yeast CST complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5XQL
 
 | | Crystal structure of a Pseudomonas aeruginosa transcriptional regulator | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Multidrug-efflux transporter 1 regulator | | Authors: | Raju, H, Sharma, R. | | Deposit date: | 2017-06-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Crystal structure of BrlR with c-di-GMP
Biochem. Biophys. Res. Commun., 490, 2017
|
|
6LBU
 
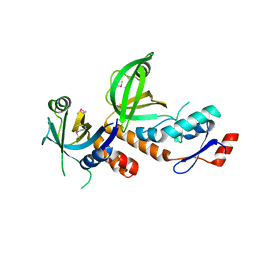 | | Crystal structure of yeast Stn1 and Ten1 | | Descriptor: | KLLA0C11825p, KLLA0E09417p | | Authors: | Ge, Y, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2019-11-14 | | Release date: | 2020-07-15 | | Last modified: | 2020-08-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into telomere protection and homeostasis regulation by yeast CST complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6LB9
 
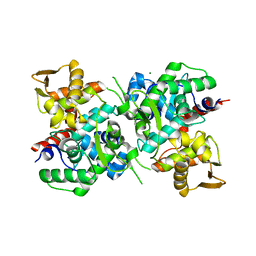 | | Magnesium ion-bound SspB crystal structure | | Descriptor: | DUF4007 domain-containing protein, MAGNESIUM ION | | Authors: | Liqiong, L, Yubing, Z. | | Deposit date: | 2019-11-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | SspABCD-SspE is a phosphorothioation-sensing bacterial defence system with broad anti-phage activities.
Nat Microbiol, 5, 2020
|
|
7QNQ
 
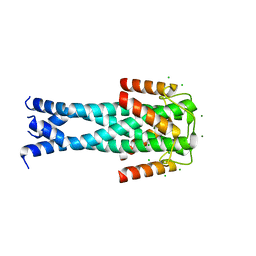 | | Structure of the Aux2 relaxosome protein of plasmid pLS20 | | Descriptor: | Auxiliary relaxosome protein, CHLORIDE ION, MAGNESIUM ION | | Authors: | Boer, D.R, Crespo, I. | | Deposit date: | 2021-12-22 | | Release date: | 2022-03-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural and biochemical characterization of the relaxosome auxiliary proteins encoded on the Bacillus subtilis plasmid pLS20.
Comput Struct Biotechnol J, 20, 2022
|
|
7QAA
 
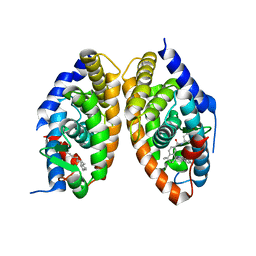 | | Crystal structure of RARalpha/RXRalpha ligand binding domain heterodimer in complex with BMS614 and oleic acid | | Descriptor: | 4-[(4,4-DIMETHYL-1,2,3,4-TETRAHYDRO-[1,2']BINAPTHALENYL-7-CARBONYL)-AMINO]-BENZOIC ACID, Isoform Alpha-1-deltaBC of Retinoic acid receptor alpha, OLEIC ACID, ... | | Authors: | le Maire, A, Vivat, V, Guee, L, Blanc, P, Malosse, C, Chamot-Rooke, J, Germain, P, Bourguet, w. | | Deposit date: | 2021-11-16 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Design and in vitro characterization of RXR variants as tools to investigate the biological role of endogenous rexinoids.
J.Mol.Endocrinol., 69, 2022
|
|
2O61
 
 | |
7YE1
 
 | | The cryo-EM structure of C. crescentus GcrA-TACup | | Descriptor: | Cell cycle regulatory protein GcrA, DNA (57-MER)-non template, DNA (57-MER)-template, ... | | Authors: | Wu, X.X, Zhang, Y. | | Deposit date: | 2022-07-05 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of Caulobacter crescentus transcription activation complex with an essential cell cycle regulator GcrA
Nucleic Acids Res., 2023
|
|
7YE2
 
 | | The cryo-EM structure of C. crescentus GcrA-TACdown | | Descriptor: | Cell cycle regulatory protein GcrA, DNA (90-MER)-non template, DNA (90-MER)-template, ... | | Authors: | Wu, X.X, Zhang, Y. | | Deposit date: | 2022-07-05 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of Caulobacter crescentus transcription activation complex with an essential cell cycle regulator GcrA
Nucleic Acids Res., 2023
|
|
6KXZ
 
 | |
5O1B
 
 | | p53 cancer mutant Y220C in complex with compound MB84 | | Descriptor: | 6-(hydroxymethyl)-2,4-bis(iodanyl)-3-pyrrol-1-yl-phenol, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Joerger, A.C, Bauer, M.R, Baud, M.G.J, Fersht, A.R. | | Deposit date: | 2017-05-18 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Aminobenzothiazole derivatives stabilize the thermolabile p53 cancer mutant Y220C and show anticancer activity in p53-Y220C cell lines.
Eur J Med Chem, 152, 2018
|
|
3F52
 
 | | Crystal structure of the clp gene regulator ClgR from C. glutamicum | | Descriptor: | GLYCEROL, clp gene regulator (ClgR) | | Authors: | Russo, S, Schweitzer, J.E, Polen, T, Bott, M, Pohl, E. | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the caseinolytic protease gene regulator, a transcriptional activator in actinomycetes
J.Biol.Chem., 284, 2009
|
|
3F51
 
 | | Crystal Structure of the clp gene regulator ClgR from Corynebacterium glutamicum | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Clp gene regulator (ClgR) | | Authors: | Russo, S, Schweitzer, J.E, Polen, T, Bott, M, Pohl, E. | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the caseinolytic protease gene regulator, a transcriptional activator in actinomycetes
J.Biol.Chem., 284, 2009
|
|
8TNQ
 
 | | Cryo-EM structure of DDB1dB:CRBN:PT-179:SD40, conformation 1 | | Descriptor: | 2-[(3S)-2,6-dioxopiperidin-3-yl]-5-(morpholin-4-yl)-1H-isoindole-1,3(2H)-dione, DNA damage-binding protein 1, Maltose/maltodextrin-binding periplasmic protein,SD40, ... | | Authors: | Roy Burman, S.S, Hunkeler, M, Fischer, E.S. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Continuous evolution of compact protein degradation tags regulated by selective molecular glues.
Science, 383, 2024
|
|
8THB
 
 | | Structure of the Saccharomyces cerevisiae PCNA clamp unloader Elg1-RFC complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ELG1 isoform 1, MAGNESIUM ION, ... | | Authors: | Zheng, F, Yao, Y.N, Georgescu, R, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-07-14 | | Release date: | 2024-05-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the PCNA unloader Elg1-RFC.
Sci Adv, 10, 2024
|
|
