8A76
 
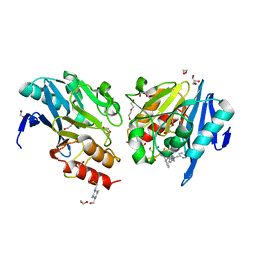 | | Metallo-beta-lactamase NDM-1 in complex with 1,2,4-Triazole-3-thione compound 26 | | Descriptor: | (2~{S})-2-[bis(pyridin-2-ylmethyl)amino]-5-(3-phenyl-5-sulfanylidene-1~{H}-1,2,4-triazol-4-yl)pentanoic acid, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Vascon, F, Legru, A, Hernandez, J.F, Cendron, L. | | Deposit date: | 2022-06-20 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Optimization of 1,2,4-Triazole-3-thiones toward Broad-Spectrum Metallo-beta-lactamase Inhibitors Showing Potent Synergistic Activity on VIM- and NDM-1-Producing Clinical Isolates.
J.Med.Chem., 65, 2022
|
|
5HPR
 
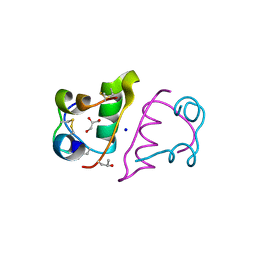 | | Insulin with proline analog HyP at position B28 in the T2 state | | Descriptor: | GLYCEROL, Insulin A-Chain, Insulin B-Chain, ... | | Authors: | Lieblich, S.A, Fang, K.Y, Cahn, J.K.B, Tirrell, D.A. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | 4S-Hydroxylation of Insulin at ProB28 Accelerates Hexamer Dissociation and Delays Fibrillation.
J. Am. Chem. Soc., 139, 2017
|
|
7US3
 
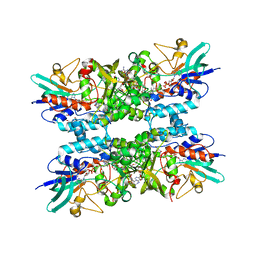 | | Structure of Putrescine N-hydroxylase Involved Complexed with NADP+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putrescine N-hydroxylase, ... | | Authors: | Tanner, J.J, Bogner, A.N. | | Deposit date: | 2022-04-22 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Kinetic and Structural Characterization of a Flavin-Dependent Putrescine N -Hydroxylase from Acinetobacter baumannii.
Biochemistry, 61, 2022
|
|
6NIE
 
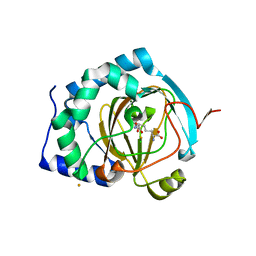 | | BesD with Fe(II), chloride, and alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, BesD, lysine halogenase, ... | | Authors: | Neugebauer, M.E, Chang, M.C.Y. | | Deposit date: | 2018-12-27 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A family of radical halogenases for the engineering of amino-acid-based products.
Nat.Chem.Biol., 15, 2019
|
|
7Q7W
 
 | |
8QRE
 
 | | Cholera holotoxin (wildtype) | | Descriptor: | ACETATE ION, Cholera enterotoxin subunit A, Cholera enterotoxin subunit B, ... | | Authors: | Mojica, N, Cordara, G, Krengel, U. | | Deposit date: | 2023-10-06 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Using Vibrio natriegens for High-Yield Production of Challenging Expression Targets and for Protein Perdeuteration.
Biochemistry, 63, 2024
|
|
8OMJ
 
 | | hKHK-C in complex with compound 28 | | Descriptor: | Ketohexokinase, SULFATE ION, [3-[[6-[(3~{a}~{R},6~{a}~{S})-2,3,3~{a},4,6,6~{a}-hexahydro-1~{H}-pyrrolo[3,4-c]pyrrol-5-yl]-3-cyano-4-(trifluoromethyl)pyridin-2-yl]amino]-4-methylsulfanyl-phenyl]methoxy-methyl-phosphinic acid | | Authors: | Pautsch, A, Ebenhoch, R. | | Deposit date: | 2023-03-31 | | Release date: | 2024-07-10 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Discovery of BI-9787, a potent zwitterionic ketohexokinase inhibitor with oral bioavailability.
Bioorg.Med.Chem.Lett., 112, 2024
|
|
7O39
 
 | |
6NIX
 
 | | Crystal structure of Immune Receptor | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, ... | | Authors: | Ting, Y.T, Reid, H.H, Rossjohn, J. | | Deposit date: | 2019-01-01 | | Release date: | 2020-01-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Immune Receptor
To Be Published
|
|
8WAE
 
 | |
5E2B
 
 | | Crystal structure of NTMT1 in complex with N-terminally methylated PPKRIA peptide | | Descriptor: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-30 | | Release date: | 2015-10-28 | | Last modified: | 2015-12-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
7UFJ
 
 | | Structure of human MR1-ethylvanillin in complex with human MAIT A-F7 TCR | | Descriptor: | 3-ethoxy-4-hydroxybenzaldehyde, ACETATE ION, Beta-2-microglobulin, ... | | Authors: | Wang, C.J, Rossjohn, J, Le Nours, J. | | Deposit date: | 2022-03-22 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Quantitative affinity measurement of small molecule ligand binding to Major Histocompatibility Complex class-I related protein 1 MR1.
J.Biol.Chem., 298, 2022
|
|
8WAI
 
 | |
7ZV8
 
 | | Crystal structure of SARS Cov-2 main protease in complex with an inhibitor 58 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, OCTANOIC ACID (CAPRYLIC ACID), ... | | Authors: | Rahimova, R, Di Micco, S, Marquez, J.A. | | Deposit date: | 2022-05-13 | | Release date: | 2022-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | Rational design of the zonulin inhibitor AT1001 derivatives as potential anti SARS-CoV-2.
Eur.J.Med.Chem., 244, 2022
|
|
6NJ4
 
 | |
5TBX
 
 | | hnRNP A18 RNA Recognition Motif | | Descriptor: | ACETATE ION, Cold-inducible RNA-binding protein, NICKEL (II) ION | | Authors: | Coburn, K.M, Melville, Z, Aligholizadeh, E, Roth, B.M, Varney, K.M, Weber, D.J. | | Deposit date: | 2016-09-13 | | Release date: | 2017-04-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.767 Å) | | Cite: | Crystal structure of the human heterogeneous ribonucleoprotein A18 RNA-recognition motif.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5HRV
 
 | | Crystal structure of the fifth bromodomain of human PB1 in complex with 1-ethylisochromeno[3,4-c]pyrazol-5(2H)-one) compound | | Descriptor: | 1,2-ETHANEDIOL, 1-ethylisochromeno[3,4-c]pyrazol-5(3H)-one, Protein polybromo-1 | | Authors: | Tallant, C, Myrianthopoulos, V, Gaboriaud-Kolar, N, Newman, J.A, Picaud, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Mikros, E, Knapp, S. | | Deposit date: | 2016-01-24 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and Optimization of a Selective Ligand for the Switch/Sucrose Nonfermenting-Related Bromodomains of Polybromo Protein-1 by the Use of Virtual Screening and Hydration Analysis.
J.Med.Chem., 59, 2016
|
|
5E3M
 
 | | Crystal structure of Fis bound to 27bp DNA F35 (AAATTAGTTTGAATCTCGAGCTAATTT) | | Descriptor: | DNA (27-MER), DNA-binding protein Fis | | Authors: | Stella, S, Hancock, S.P, Cascio, D, Johnson, R.C. | | Deposit date: | 2015-10-03 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.886 Å) | | Cite: | DNA Sequence Determinants Controlling Affinity, Stability and Shape of DNA Complexes Bound by the Nucleoid Protein Fis.
Plos One, 11, 2016
|
|
6NJT
 
 | | Mouse endonuclease G mutant - H97A | | Descriptor: | CHLORIDE ION, Endonuclease G, mitochondrial, ... | | Authors: | Vander Zanden, C.M, Ho, E.N, Czarny, R.S, Robertson, A.B, Ho, P.S. | | Deposit date: | 2019-01-04 | | Release date: | 2020-01-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural adaptation of vertebrate endonuclease G for 5-hydroxymethylcytosine recognition and function.
Nucleic Acids Res., 48, 2020
|
|
7UMW
 
 | | Crystal structure of E. Coli FabI in complex with NAD and Fabimycin ((S,E)-3-(7-amino-8-oxo-6,7,8,9-tetrahydro-5H-pyrido[2,3-b]azepin-3-yl)-N-methyl-N-((3-methylbenzofuran-2-yl)methyl)acrylamide) | | Descriptor: | (2E)-3-[(7S)-7-amino-8-oxo-6,7,8,9-tetrahydro-5H-pyrido[2,3-b]azepin-3-yl]-N-methyl-N-[(3-methyl-1-benzofuran-2-yl)methyl]prop-2-enamide, Enoyl-[acyl-carrier-protein] reductase [NADH] FabI, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hajian, B. | | Deposit date: | 2022-04-08 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | An Iterative Approach Guides Discovery of the FabI Inhibitor Fabimycin, a Late-Stage Antibiotic Candidate with In Vivo Efficacy against Drug-Resistant Gram-Negative Infections
Acs Cent.Sci., 8, 2022
|
|
8QZ1
 
 | | Crystal structure of human two pore domain potassium ion channel TREK-2 (K2P10.1) in complex with a nanobody (Nb58) | | Descriptor: | Isoform B of Potassium channel subfamily K member 10, Nanobody 58, POTASSIUM ION | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Baronina, A, Ang, J, Bushell, S.R, Chalk, R, Mukhopadhyay, S.M.M, Pardon, E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Tucker, S.J, Steyaert, J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.588 Å) | | Cite: | Extracellular modulation of TREK-2 activity with nanobodies provides insight into the mechanisms of K2P channel regulation.
Nat Commun, 15, 2024
|
|
5T5U
 
 | |
8QZ3
 
 | | Crystal structure of human two pore domain potassium ion channel TREK-2 (K2P10.1) in complex with an activatory nanobody (Nb67) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Nanobody 67, POTASSIUM ION, ... | | Authors: | Baronina, A, Pike, A.C.W, Rodstrom, K.E.J, Ang, J, Bushell, S.R, Chalk, R, Mukhopadhyay, S.M.M, Pardon, E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Tucker, S.J, Steyaert, J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Extracellular modulation of TREK-2 activity with nanobodies provides insight into the mechanisms of K2P channel regulation.
Nat Commun, 15, 2024
|
|
5E40
 
 | | 3-Deoxy-D-arabino-heptulosonate 7-phosphate synthase from Mycobacterium tuberculosis with D-tyrosine bound in the phenylalanine binding site | | Descriptor: | 3-deoxy-D-arabinoheptulosonate-7-phosphate synthase, D-TYROSINE, GLYCEROL, ... | | Authors: | Reichau, S, Jiao, W, Parker, E.J. | | Deposit date: | 2015-10-05 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Probing the Sophisticated Synergistic Allosteric Regulation of Aromatic Amino Acid Biosynthesis in Mycobacterium tuberculosis Using -Amino Acids.
Plos One, 11, 2016
|
|
7ZR3
 
 | |
