7RT9
 
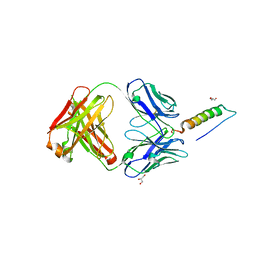 | | Crystal structures of human PYY and NPY | | Descriptor: | 4A3B2-A Fab heavy chain, 4A3B2-A Fab light chain, GLYCEROL, ... | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2021-08-12 | | Release date: | 2022-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of human neuropeptide Y (NPY) and peptide YY (PYY).
Neuropeptides, 92, 2022
|
|
3Q68
 
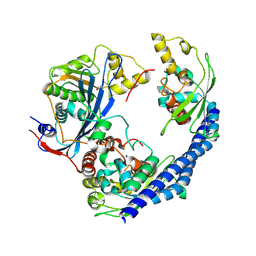 | |
7RTA
 
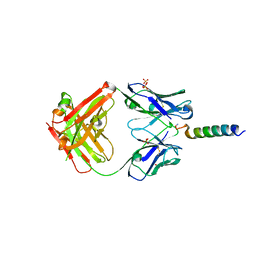 | | Crystal structures of human PYY and NPY | | Descriptor: | 4A3B2-B Fab heavy chain, 4A3B2-B Fab light chain, Neuropeptide Y, ... | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2021-08-12 | | Release date: | 2022-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of human neuropeptide Y (NPY) and peptide YY (PYY).
Neuropeptides, 92, 2022
|
|
7OC1
 
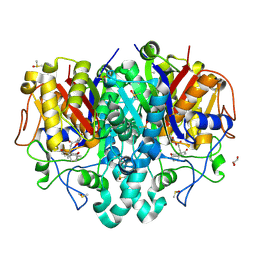 | | Structure of Pseudomonas aeruginosa FabF mutant C164Q in complex with Platensimycin | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Georgiou, C, Brenk, R, Espeland, L.O, Klein, R. | | Deposit date: | 2021-04-25 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Experimental Toolbox for Structure-Based Hit Discovery for P. aeruginosa FabF, a Promising Target for Antibiotics.
Chemmedchem, 16, 2021
|
|
3QOR
 
 | |
7OC0
 
 | | Structure of Pseudomonas aeruginosa FabF mutant C164Q in complex with a ligand (2S,4R)-2-(thiophen-2-yl)thiazolidine-4-carboxylic acid | | Descriptor: | (2S,4R)-2-(thiophen-2-yl)thiazolidine-4-carboxylic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Georgiou, C, Brenk, R, Espeland, L.O, Klein, R. | | Deposit date: | 2021-04-25 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | An Experimental Toolbox for Structure-Based Hit Discovery for P. aeruginosa FabF, a Promising Target for Antibiotics.
Chemmedchem, 16, 2021
|
|
7O6N
 
 | | Crystal structure of C. elegans ERH-2 PID-3 complex | | Descriptor: | Enhancer of rudimentary homolog 2, FORMIC ACID, Protein pid-3 | | Authors: | Falk, S, Ketting, R.F. | | Deposit date: | 2021-04-11 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural basis of PETISCO complex assembly during piRNA biogenesis in C. elegans .
Genes Dev., 35, 2021
|
|
7OCX
 
 | |
7OCZ
 
 | |
7NT7
 
 | |
7O6L
 
 | | Crystal structure of C. elegans ERH-2 | | Descriptor: | Enhancer of rudimentary homolog 2 | | Authors: | Falk, S, Ketting, R.F. | | Deposit date: | 2021-04-11 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of PETISCO complex assembly during piRNA biogenesis in C. elegans .
Genes Dev., 35, 2021
|
|
7PCJ
 
 | | X-ray structure of CypA-C52AK125C/CsA/aromatic foldamer complex | | Descriptor: | 8-azanyl-4-(2-methylpropoxy)quinoline-2-carboxylic acid, 8-azanyl-4-(3-azanylpropoxy)quinoline-2-carboxylic acid, 8-azanyl-4-oxidanyl-quinoline-2-carboxylic acid, ... | | Authors: | Vallade, M, Langlois d'Estaintot, B, Fischer, L, Buratto, J, Savko, M, Huc, I. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | X-ray structure of a cystein mutant of Cyclophilin A tethered to an aromatic oligoamide foldamer complexed with Cyclosporin A
To Be Published
|
|
3S7C
 
 | | Crystal structure of a 2'-azido-uridine-modified RNA | | Descriptor: | Ecoli 23 S rRNA Sarcin Ricin loop | | Authors: | Ennifar, E, Micura, R. | | Deposit date: | 2011-05-26 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | 2'-Azido RNA, a Versatile Tool for Chemical Biology: Synthesis, X-ray Structure, siRNA Applications, Click Labeling.
Acs Chem.Biol., 7, 2012
|
|
7VEI
 
 | | Neutron structure of D2O-solvent lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, NICKEL (II) ION | | Authors: | Chatake, T, Tanaka, I, Kusaka, K, Fujiwara, S. | | Deposit date: | 2021-09-08 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | NEUTRON DIFFRACTION (2 Å) | | Cite: | Protonation states of hen egg-white lysozyme observed using D/H contrast neutron crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7RIB
 
 | | Griffithsin mutant Y28F/Y68F/Y110F | | Descriptor: | Griffithsin, SULFATE ION, alpha-D-mannopyranose | | Authors: | Zhao, G, Sun, J, Bewley, C.A. | | Deposit date: | 2021-07-19 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | C 3 -Symmetric Aromatic Core of Griffithsin Is Essential for Potent Anti-HIV Activity.
Acs Chem.Biol., 17, 2022
|
|
7RKI
 
 | | Griffithsin-S10Y/S42Y/S88Y | | Descriptor: | Griffithsin, alpha-D-mannopyranose | | Authors: | Sun, J.D, Zhao, G.X. | | Deposit date: | 2021-07-22 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | C 3 -Symmetric Aromatic Core of Griffithsin Is Essential for Potent Anti-HIV Activity.
Acs Chem.Biol., 17, 2022
|
|
7RIA
 
 | | Griffithsin variant Y28A/Y68A/Y110A | | Descriptor: | Griffithsin, SULFATE ION, alpha-D-mannopyranose | | Authors: | Zhao, G, Sun, J, Bewley, C.A. | | Deposit date: | 2021-07-19 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C 3 -Symmetric Aromatic Core of Griffithsin Is Essential for Potent Anti-HIV Activity.
Acs Chem.Biol., 17, 2022
|
|
7RIC
 
 | | Griffithsin variant Y28W/Y68W/Y110W | | Descriptor: | Griffithsin | | Authors: | Zhao, G, Sun, J, Bewley, C.A. | | Deposit date: | 2021-07-19 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | C 3 -Symmetric Aromatic Core of Griffithsin Is Essential for Potent Anti-HIV Activity.
Acs Chem.Biol., 17, 2022
|
|
7RID
 
 | | Griffithsin variant Y28A | | Descriptor: | Griffithsin | | Authors: | Zhao, G, Sun, J, Bewley, C.A. | | Deposit date: | 2021-07-19 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | C 3 -Symmetric Aromatic Core of Griffithsin Is Essential for Potent Anti-HIV Activity.
Acs Chem.Biol., 17, 2022
|
|
7RKG
 
 | | Griffithsin mutant Y28W | | Descriptor: | Griffithsin, alpha-D-mannopyranose | | Authors: | Zhao, G, Sun, J, Bewley, C. | | Deposit date: | 2021-07-22 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | C 3 -Symmetric Aromatic Core of Griffithsin Is Essential for Potent Anti-HIV Activity.
Acs Chem.Biol., 17, 2022
|
|
7VE4
 
 | | C-terminal domain of VraR | | Descriptor: | DNA-binding response regulator | | Authors: | Kumar, J.V, Chen, C, Hsu, C.H. | | Deposit date: | 2021-09-08 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural insights into DNA binding domain of vancomycin-resistance-associated response regulator in complex with its promoter DNA from Staphylococcus aureus.
Protein Sci., 31, 2022
|
|
7PZI
 
 | |
7PZ9
 
 | | HBc-F97L premature secretion phenotype | | Descriptor: | Capsid protein | | Authors: | Makbul, C, Boettcher, B. | | Deposit date: | 2021-10-11 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Binding of a Pocket Factor to Hepatitis B Virus Capsids Changes the Rotamer Conformation of Phenylalanine 97.
Viruses, 13, 2021
|
|
7PZK
 
 | | HBc-WT in complex with Triton X-100 | | Descriptor: | Capsid protein, FRAGMENT OF TRITON X-100 | | Authors: | Makbul, C, Boettcher, B. | | Deposit date: | 2021-10-12 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Binding of a Pocket Factor to Hepatitis B Virus Capsids Changes the Rotamer Conformation of Phenylalanine 97.
Viruses, 13, 2021
|
|
7PZM
 
 | | HBc-P5T in complex with X-100 | | Descriptor: | Capsid protein, FRAGMENT OF TRITON X-100 | | Authors: | Makbul, C, Boettcher, B. | | Deposit date: | 2021-10-12 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Binding of a Pocket Factor to Hepatitis B Virus Capsids Changes the Rotamer Conformation of Phenylalanine 97.
Viruses, 13, 2021
|
|
