2LMD
 
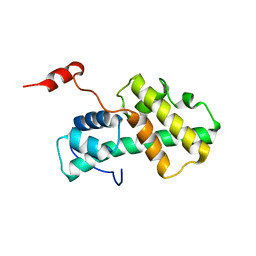 | | Minimal Constraints Solution NMR Structure of Prospero Homeobox protein 1 from Homo sapiens, Northeast Structural Genomics Consortium Target HR4660B | | Descriptor: | Prospero homeobox protein 1 | | Authors: | Rossi, P, Lange, O.A, Lee, H, Maglaqui, M, Janjua, H, Ciccosanti, C, Zhao, L, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-11-29 | | Release date: | 2011-12-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2JZN
 
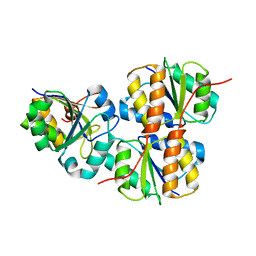 | |
2JVW
 
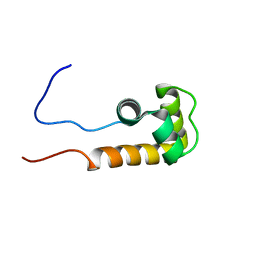 | | Solution NMR structure of uncharacterized protein Q5E7H1 from Vibrio fischeri. Northeast Structural Genomics target VfR117 | | Descriptor: | Uncharacterized protein | | Authors: | Aramini, J.M, Rossi, P, Wang, D, Nwosu, C, Owens, L.A, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-26 | | Release date: | 2007-10-16 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of VF0530 from Vibrio fischeri reveals a nucleic acid-binding function.
Proteins, 79, 2011
|
|
2H49
 
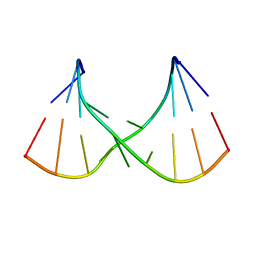 | |
2L0P
 
 | | Solution structure of human apo-S100A1 protein by NMR spectroscopy | | Descriptor: | S100 calcium binding protein A1 | | Authors: | Nowakowski, M, Jaremko, L, Jaremko, M, Bierzynski, A, Zhukov, I, Ejchart, A. | | Deposit date: | 2010-07-12 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure and dynamics of human apo-S100A1 protein.
J.Struct.Biol., 174, 2011
|
|
2JZO
 
 | |
1WRF
 
 | | Refined solution structure of Der f 2, The Major Mite Allergen from Dermatophagoides farinae | | Descriptor: | Mite group 2 allergen Der f 2 | | Authors: | Ichikawa, S, Takai, T, Inoue, T, Yuuki, T, Okumura, Y, Ogura, K, Inagaki, F, Hatanaka, H. | | Deposit date: | 2004-10-15 | | Release date: | 2005-04-19 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR Study on the Major Mite Allergen Der f 2: Its Refined Tertiary Structure, Epitopes for Monoclonal Antibodies and Characteristics Shared by ML Protein Group Members
J.Biochem.(Tokyo), 137, 2005
|
|
2JOB
 
 | | Solution structure of an antilipopolysaccharide factor from shrimp and its possible Lipid A binding site | | Descriptor: | antilipopolysaccharide factor | | Authors: | Yang, Y, Boze, H, Chemardin, P, Padilla, A, Moulin, G, Tassanakajon, A, Pugniere, M, Roquet, F, Gueguen, Y, Bachere, E, Aumelas, A. | | Deposit date: | 2007-03-02 | | Release date: | 2008-03-11 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | NMR structure of rALF-Pm3, an anti-lipopolysaccharide factor from shrimp: Model of the possible lipid A-binding site
Biopolymers, 91, 2009
|
|
2KRT
 
 | | Solution NMR Structure of a Conserved Hypothetical Membrane Lipoprotein Obtained from Ureaplasma parvum: Northeast Structural Genomics Consortium Target UuR17A (139-239) | | Descriptor: | Conserved hypothetical membrane lipoprotein | | Authors: | Mani, R, Swapna, G, Janjua, H, Ciccosanti, C, Huang, Y, Patel, D, Xiao, R, Acton, T, Everett, J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-12-22 | | Release date: | 2010-01-05 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of a Conserved Hypothetical Membrane Lipoprotein obtained from Ureaplasma parvum : Northeast Structural Genomics Consortium Target UuR17A (139-239)
To be Published
|
|
2KRJ
 
 | | High-Resolution Solid-State NMR Structure of a 17.6 kDa Protein | | Descriptor: | COBALT (II) ION, Macrophage metalloelastase | | Authors: | Bertini, I, Bhaumik, A, De Pa pe, G, Griffin, R.G, Lelli, M, Lewandowski, J.R, Luchinat, C. | | Deposit date: | 2009-12-18 | | Release date: | 2010-02-23 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution solid-state NMR structure of a 17.6 kDa protein.
J.Am.Chem.Soc., 132, 2010
|
|
2KSB
 
 | |
2KSA
 
 | |
2L6Q
 
 | |
2DAU
 
 | | DICKERSON-DREW DNA DODECAMER, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Denisov, A, Bekiroglu, S, Maltseva, T, Sandstrom, A, Altmann, K.-H, Egli, M, Chattopadhyaya, J. | | Deposit date: | 1998-01-20 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution conformation of a carbocyclic analog of the Dickerson-Drew dodecamer: comparison with its own X-ray structure and that of the NMR structure of the native counterpart.
J.Biomol.Struct.Dyn., 16, 1998
|
|
2KPP
 
 | | Solution NMR structure of Lin0431 protein from Listeria innocua. Northeast Structural Genomics Consortium Target LkR112 | | Descriptor: | Lin0431 protein | | Authors: | Tang, Y, Xiao, R, Ciccosanti, C, Janjua, H, Lee, D.Y, Everett, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-18 | | Release date: | 2010-02-23 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Lin0431 protein from Listeria innocua reveals high structural similarity with domain II of bacterial transcription antitermination protein NusG.
Proteins, 78, 2010
|
|
2EZA
 
 | | AMINO TERMINAL DOMAIN OF ENZYME I FROM ESCHERICHIA COLI, NMR, RESTRAINED REGULARIZED MEAN STRUCTURE | | Descriptor: | PHOSPHOTRANSFERASE SYSTEM, ENZYME I | | Authors: | Clore, G.M, Tjandra, N, Garrett, D.S, Gronenborn, A.M. | | Deposit date: | 1997-05-07 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Defining long range order in NMR structure determination from the dependence of heteronuclear relaxation times on rotational diffusion anisotropy.
Nat.Struct.Biol., 4, 1997
|
|
2EZC
 
 | | AMINO TERMINAL DOMAIN OF ENZYME I FROM ESCHERICHIA COLI, NMR, 14 STRUCTURES | | Descriptor: | PHOSPHOTRANSFERASE SYSTEM, ENZYME I | | Authors: | Clore, G.M, Tjandra, N, Garrett, D.S, Gronenborn, A.M. | | Deposit date: | 1997-05-07 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Defining long range order in NMR structure determination from the dependence of heteronuclear relaxation times on rotational diffusion anisotropy.
Nat.Struct.Biol., 4, 1997
|
|
2EZB
 
 | | AMINO TERMINAL DOMAIN OF ENZYME I FROM ESCHERICHIA COLI, NMR, 14 STRUCTURES | | Descriptor: | PHOSPHOTRANSFERASE SYSTEM, ENZYME I | | Authors: | Clore, G.M, Tjandra, N, Garrett, D.S, Gronenborn, A.M. | | Deposit date: | 1997-05-07 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Defining long range order in NMR structure determination from the dependence of heteronuclear relaxation times on rotational diffusion anisotropy.
Nat.Struct.Biol., 4, 1997
|
|
1WLP
 
 | | Solution Structure Of The P22Phox-P47Phox Complex | | Descriptor: | Cytochrome b-245 light chain, Neutrophil cytosol factor 1 | | Authors: | Ogura, K, Torikai, S, Saikawa, K, Yuzawa, S, Sumimoto, H, Inagaki, F. | | Deposit date: | 2004-06-29 | | Release date: | 2005-10-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the tandem Src homology 3 domains of p47phox complexed with a p22phox-derived proline-rich peptide
J.Biol.Chem., 281, 2006
|
|
2C0X
 
 | | MOLECULAR STRUCTURE OF FD FILAMENTOUS BACTERIOPHAGE REFINED WITH RESPECT TO X-RAY FIBRE DIFFRACTION AND SOLID-STATE NMR DATA | | Descriptor: | COAT PROTEIN B | | Authors: | Marvin, D.A, Welsh, L.C, Symmons, M.F, Scott, W.R.P, Straus, S.K. | | Deposit date: | 2005-09-08 | | Release date: | 2005-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Molecular Structure of Fd (F1, M13) Filamentous Bacteriophage Refined with Respect to X-Ray Fibre Diffraction and Solid-State NMR Data Supports Specific Models of Phage Assembly at the Bacterial Membrane.
J.Mol.Biol., 355, 2006
|
|
2BVK
 
 | |
2K56
 
 | |
2KJ7
 
 | | Three-Dimensional NMR Structure of Rat Islet Amyloid Polypeptide in DPC micelles | | Descriptor: | Islet amyloid polypeptide | | Authors: | Nanga, R, Brender, J.R, Xu, J, Hartman, K, Subramanian, V, Ramamoorthy, A. | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-23 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure and orientation of rat islet amyloid polypeptide protein in a membrane environment by solution NMR spectroscopy.
J.Am.Chem.Soc., 131, 2009
|
|
1Z65
 
 | | Mouse Doppel 1-30 peptide | | Descriptor: | Prion-like protein doppel | | Authors: | Papadopoulos, E. | | Deposit date: | 2005-03-21 | | Release date: | 2006-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Peptide Fragment 1-30, Derived from Unprocessed Mouse Doppel Protein, in DHPC Micelles
Biochemistry, 45, 2006
|
|
2KOD
 
 | | A high-resolution NMR structure of the dimeric C-terminal domain of HIV-1 CA | | Descriptor: | HIV-1 CA C-terminal domain | | Authors: | Byeon, I.-J.L, Jung, J, Ahn, J, concel, J, Gronenborn, A.M. | | Deposit date: | 2009-09-18 | | Release date: | 2009-11-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural convergence between Cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function.
Cell(Cambridge,Mass.), 139, 2009
|
|
