4XHF
 
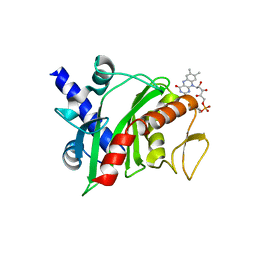 | | Crystal structure of Shewanella oneidensis NqrC | | Descriptor: | FLAVIN MONONUCLEOTIDE, Na-translocating NADH-quinone reductase subunit C NqrC, SODIUM ION | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2015-01-05 | | Release date: | 2015-12-16 | | Last modified: | 2016-03-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular insights into the enzymatic diversity of flavin-trafficking protein (Ftp; formerly ApbE) in flavoprotein biogenesis in the bacterial periplasm.
Microbiologyopen, 5, 2016
|
|
8P41
 
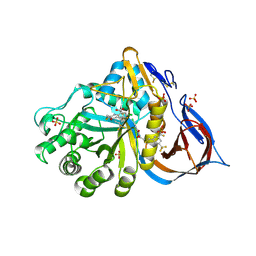 | | Crystal structure of glucocerebrosidase in complex with allosteric activator | | Descriptor: | 2-[2-[[3,5-bis(trifluoromethyl)phenyl]methylsulfanyl]ethanoylamino]-5-chloranyl-benzoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Schulze, M.-S. | | Deposit date: | 2023-05-18 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Identification of ss-Glucocerebrosidase Activators for Glucosylceramide hydrolysis.
Chemmedchem, 19, 2024
|
|
4XIX
 
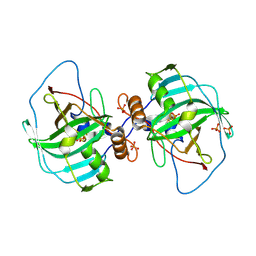 | | Carbonic anhydrase Cah3 from Chlamydomonas reinhardtii in complex with phosphate. | | Descriptor: | Carbonic anhydrase, alpha type, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Hainzl, T, Grundstrom, C, Benlloch, R, Shevela, D, Shutova, T, Messinger, J, Samuelsson, G, Sauer-Eriksson, A.E. | | Deposit date: | 2015-01-08 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure and Functional Characterization of Photosystem II-Associated Carbonic Anhydrase CAH3 in Chlamydomonas reinhardtii.
Plant Physiol., 167, 2015
|
|
8PAY
 
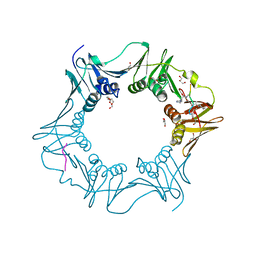 | | Structure of the E.coli DNA polymerase sliding clamp with a covalently bound peptide 2. | | Descriptor: | ACE-GLN-ALC-GLC-LEU-PHE, Beta sliding clamp, GLYCEROL, ... | | Authors: | Compain, G, Monsarrat, C, Blagojevic, J, Brillet, K, Dumas, P, Hammann, P, Kuhn, L, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Burnouf, D, wagner, J, Guichard, G. | | Deposit date: | 2023-06-08 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Peptide-Based Covalent Inhibitors Bearing Mild Electrophiles to Target a Conserved His Residue of the Bacterial Sliding Clamp.
Jacs Au, 4, 2024
|
|
8UR3
 
 | |
8UR6
 
 | |
8U3N
 
 | |
8UKZ
 
 | |
8U1D
 
 | |
8U2M
 
 | |
7NMB
 
 | |
4XJB
 
 | | X-ray structure of Lysozyme1 | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XJG
 
 | | X-ray structure of Lysozyme B2 | | Descriptor: | BROMIDE ION, Lysozyme C, SODIUM ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-06-03 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XJI
 
 | | X-ray structure of LysozymeS2 | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-06-03 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8PAT
 
 | | Structure of the E.coli DNA polymerase sliding clamp with a covalently bound peptide 3. | | Descriptor: | ACE-GLN-ALC-GLX-LEU-PHE, Beta sliding clamp | | Authors: | Compain, G, Monsarrat, C, Blagojevic, J, Brillet, K, Dumas, P, Hammann, P, Kuhn, L, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Burnouf, D, Guichard, G. | | Deposit date: | 2023-06-08 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Peptide-Based Covalent Inhibitors Bearing Mild Electrophiles to Target a Conserved His Residue of the Bacterial Sliding Clamp.
Jacs Au, 4, 2024
|
|
8V44
 
 | |
7N03
 
 | | Crystal structure of MTH1 in complex with compound 31 | | Descriptor: | 4-anilino-6-(hexylamino)-N-methylquinoline-3-carboxamide, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Eron, S.J. | | Deposit date: | 2021-05-24 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Development of an AchillesTAG degradation system and its application to control CAR-T activity
Curr Res Chem Biol, 1, 2021
|
|
1FN7
 
 | | COUPLING OF DAMAGE RECOGNITION AND CATALYSIS BY A HUMAN BASE-EXCISION DNA REPAIR PROTEIN | | Descriptor: | 8-OXOGUANINE DNA GLYCOSYLASE 1, CALCIUM ION, DNA (5'-D(*GP*CP*GP*TP*CP*CP*AP*(3DR)P*GP*TP*CP*TP*AP*CP*C)-3'), ... | | Authors: | Norman, D.P.G, Bruner, S.D, Verdine, G.L. | | Deposit date: | 2000-08-21 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Coupling of substrate recognition and catalysis by a human base-excision DNA repair protein.
J.Am.Chem.Soc., 123, 2001
|
|
4Y1P
 
 | | Crystal structure of 3-isopropylmalate dehydrogenase (Saci_0600) from Sulfolobus acidocaldarius complex with 3-isopropylmalate and Mg2+ | | Descriptor: | 3-ISOPROPYLMALIC ACID, 3-isopropylmalate dehydrogenase, MAGNESIUM ION, ... | | Authors: | Takahashi, K, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2015-02-08 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of two beta-decarboxylating dehydrogenases from Sulfolobus acidocaldarius
Extremophiles, 20, 2016
|
|
7N13
 
 | | Crystal structure of MTH1 in complex with compound 32 | | Descriptor: | 4-anilino-6-[4-(butylcarbamoyl)-3-fluorophenyl]-N-cyclopropyl-7-fluoroquinoline-3-carboxamide, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Eron, S.J. | | Deposit date: | 2021-05-26 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Development of an AchillesTAG degradation system and its application to control CAR-T activity
Curr Res Chem Biol, 1, 2021
|
|
4XQD
 
 | | X-ray structure analysis of xylanase-WT at pH4.0 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-19 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XAZ
 
 | | Cycles of destabilization and repair underlie evolutionary transitions in enzymes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Phosphotriesterase variant PTE-R18, ZINC ION | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-16 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4XD4
 
 | | Phosphotriesterase variant E2b | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-R3, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-19 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4XES
 
 | | Structure of active-like neurotensin receptor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Krumm, B.E, White, J.F, Shah, P, Grisshammer, R. | | Deposit date: | 2014-12-24 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural prerequisites for G-protein activation by the neurotensin receptor.
Nat Commun, 6, 2015
|
|
4XJD
 
 | | X-ray structure of Lysozyme2 | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
