1GYN
 
 | | Class II fructose 1,6-bisphosphate aldolase with Cadmium (not Zinc) in the active site | | Descriptor: | CADMIUM ION, FRUCTOSE-BISPHOSPHATE ALDOLASE II | | Authors: | Hall, D.R, Kemp, L.E, Leonard, G.A, Berry, A, Hunter, W.N. | | Deposit date: | 2002-04-27 | | Release date: | 2003-02-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Organization of Divalent Cations in the Active Site of Cadmium Escherichia Coli Fructose 1,6-Bisphosphate Aldolase
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1GQ4
 
 | |
1GQM
 
 | | The structure of S100A12 in a hexameric form and its proposed role in receptor signalling | | Descriptor: | CALCIUM ION, CALGRANULIN C | | Authors: | Moroz, O.V, Antson, A.A, Dodson, E.G, Burrel, H.J, Grist, S.J, Lloyd, R.M, Maitland, N.J, Dodson, G.G, Wilson, K.S, Lukanidin, E, Bronstein, I.B. | | Deposit date: | 2001-11-26 | | Release date: | 2002-02-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of S100A12 in a Hexameric Form and its Proposed Role in Receptor Signalling
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1GSM
 
 | | A reassessment of the MAdCAM-1 structure and its role in integrin recognition. | | Descriptor: | MUCOSAL ADDRESSIN CELL ADHESION MOLECULE-1 | | Authors: | Dando, J, Wilkinson, K.W, Ortlepp, S, King, D.J, Brady, R.L. | | Deposit date: | 2002-01-08 | | Release date: | 2002-01-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Reassessment of the Madcam-1 Structure and its Role in Integrin Recognition
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4WVS
 
 | | Crystal structure of XIAP-BIR2 domain complexed with (S)-3-(4-methoxyphenyl)-2-((S)-2-((S)-1-((S)-2-((S)-2-(methylamino)propanamido)pent-4-ynoyl)pyrrolidine-2-carboxamido)-3-phenylpropanamido)propanoic acid | | Descriptor: | 3,11-DIFLUORO-6,8,13-TRIMETHYL-8H-QUINO[4,3,2-KL]ACRIDIN-13-IUM, E3 ubiquitin-protein ligase XIAP, GLYCEROL, ... | | Authors: | Pokross, M.E. | | Deposit date: | 2014-11-07 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The Discovery of Macrocyclic XIAP Antagonists from a DNA-Programmed Chemistry Library, and Their Optimization To Give Lead Compounds with in Vivo Antitumor Activity.
J.Med.Chem., 58, 2015
|
|
1GTK
 
 | | Time-resolved and static-ensemble structural chemistry of hydroxymethylbilane synthase | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, PORPHOBILINOGEN DEAMINASE | | Authors: | Helliwell, J.R, Nieh, Y.P, Raftery, J, Cassetta, A, Habash, J, Carr, P.D, Ursby, T, Wulff, M, Thompson, A.W, Niemann, A.C, Haedener, A. | | Deposit date: | 2002-01-16 | | Release date: | 2003-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Time-Resolved and Static-Ensemble Structural Chemistry of Hydroxymethylbilane Synthase
Faraday Discuss., 122, 2003
|
|
1GUS
 
 | |
6YRP
 
 | | Crystal Structure of the VIM-2 Acquired Metallo-beta-Lactamase in Complex with JMV-4690 (Cpd 31) | | Descriptor: | 1,2-ETHANEDIOL, 2-[[[3-(5-methoxy-2-oxidanyl-phenyl)-5-sulfanylidene-1~{H}-1,2,4-triazol-4-yl]amino]methyl]benzoic acid, ACETATE ION, ... | | Authors: | Docquier, J.D, Pozzi, C, De Luca, F, Benvenuti, M, Mangani, S. | | Deposit date: | 2020-04-20 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 4-Amino-1,2,4-triazole-3-thione-derived Schiff bases as metallo-beta-lactamase inhibitors.
Eur.J.Med.Chem., 208, 2020
|
|
8FO4
 
 | |
8FUJ
 
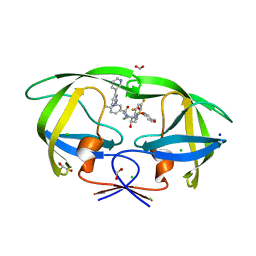 | | HIV-1 wild type protease with GRL-03419A, with N-(2,5-dimethylphenyl)-4-(pyridin-3-yl)pyrimidin-2-amine as P2-P3 group and 3,5-difluorophenylmethyl as the P1 group | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2023-01-17 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Exploration of imatinib and nilotinib-derived templates as the P2-Ligand for HIV-1 protease inhibitors: Design, synthesis, protein X-ray structural studies, and biological evaluation.
Eur.J.Med.Chem., 255, 2023
|
|
1GY9
 
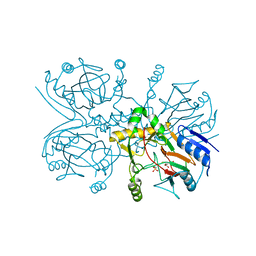 | | Taurine/alpha-ketoglutarate Dioxygenase from Escherichia coli | | Descriptor: | 2-AMINOETHANESULFONIC ACID, 2-OXOGLUTARIC ACID, ALPHA-KETOGLUTARATE-DEPENDENT TAURINE DIOXYGENASE, ... | | Authors: | Elkins, J.M, Burzlaff, N.I, Ryle, M.J, Lloyd, J.S, Clifton, I.J, Baldwin, J.E, Hausinger, R.P, Roach, P.L. | | Deposit date: | 2002-04-22 | | Release date: | 2002-04-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray Crystal Structure of Escherichia Coli Taurine/Alpha-Ketoglutarate Dioxygenase Complexed to Ferrous Iron and Substrates.
Biochemistry, 41, 2002
|
|
1GZ8
 
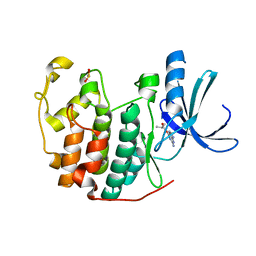 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR 2-Amino-6-(3'-methyl-2'-oxo)butoxypurine | | Descriptor: | 1-[(2-AMINO-6,9-DIHYDRO-1H-PURIN-6-YL)OXY]-3-METHYL-2-BUTANOL, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Davies, T, Endicott, J, Johnson, L, Noble, M, Tucker, J. | | Deposit date: | 2002-05-17 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Probing the ATP Ribose-Binding Domain of Cyclin-Dependent Kinases 1 and 2 with O(6)-Substituted Guanine Derivatives
J.Med.Chem., 45, 2002
|
|
1H4D
 
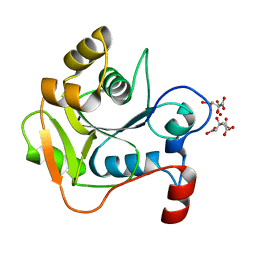 | | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis protein MobA | | Descriptor: | CITRIC ACID, LITHIUM ION, MOLYBDOPTERIN-GUANINE DINUCLEOTIDE BIOSYNTHESIS PROTEIN A | | Authors: | Guse, A, Stevenson, C.E.M, Kuper, J, Buchanan, G, Schwarz, G, Mendel, R.R, Lawson, D.M, Palmer, T. | | Deposit date: | 2003-02-26 | | Release date: | 2003-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis Protein Moba
J.Biol.Chem., 278, 2003
|
|
1H0R
 
 | | Type II Dehydroquinase from Mycobacterium tuberculosis complexed with 2,3-anhydro-quinic acid | | Descriptor: | 2,3 -ANHYDRO-QUINIC ACID, 3-DEHYDROQUINATE DEHYDRATASE, CHLORIDE ION, ... | | Authors: | Roszak, A.W, Robinson, D.A, Frederickson, M, Abell, C, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2002-06-27 | | Release date: | 2003-10-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Selectivity of Oxime Based Inhibitors Towards Type II Dehydroquinase from Mycobacterium Tuberculosis
To be Published
|
|
2DUT
 
 | |
1H0K
 
 | |
6YRW
 
 | | SHMT from Streptococcus thermophilus Tyr55Ser variant as internal aldimine and as non-covalent complex with D-Ser | | Descriptor: | D-SERINE, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Petrillo, G, Hernandez, K, Bujons, J, Clapes, P, Uson, I. | | Deposit date: | 2020-04-20 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into nucleophile substrate specificity in variants of N-Serine hydroxymethyltransferase from Streptococcus thermophilus
To Be Published
|
|
8FIV
 
 | |
1H0H
 
 | | Tungsten containing Formate Dehydrogenase from Desulfovibrio Gigas | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Raaijmakers, H.C.A. | | Deposit date: | 2002-06-19 | | Release date: | 2003-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gene Sequence and the 1.8 A Crystal Structure of the Tungsten-Containing Formate Dehydrogenase from Desulfovibrio Gigas
Structure, 10, 2002
|
|
1GXD
 
 | | proMMP-2/TIMP-2 complex | | Descriptor: | 72 KDA TYPE IV COLLAGENASE, CALCIUM ION, METALLOPROTEINASE INHIBITOR 2, ... | | Authors: | Morgunova, E, Tuuttila, A, Bergmann, U, Tryggvason, K. | | Deposit date: | 2002-04-02 | | Release date: | 2002-07-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Insight Into the Complex Formation of Latent Matrix Metalloproteinase 2 with Tissue Inhibitor of Metalloproteinase 2
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
8FIW
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor Jun10221 | | Descriptor: | 3C-like proteinase nsp5, N-([1,1'-biphenyl]-4-yl)-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]prop-2-enamide, N-([1,1'-biphenyl]-4-yl)-N-[(1S)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]prop-2-enamide | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2022-12-16 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Exploring diverse reactive warheads for the design of SARS-CoV-2 main protease inhibitors.
Eur.J.Med.Chem., 259, 2023
|
|
1HD5
 
 | |
2DGN
 
 | | Mouse Muscle Adenylosuccinate Synthetase partially ligated complex with GTP, 2'-deoxy-IMP | | Descriptor: | 9-(2-DEOXY-5-O-PHOSPHONO-BETA-D-ERYTHRO-PENTOFURANOSYL)-6-(PHOSPHONOOXY)-9H-PURINE, Adenylosuccinate synthetase isozyme 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Iancu, C.V, Zhou, Y, Borza, T, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2006-03-15 | | Release date: | 2006-09-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cavitation as a mechanism of substrate discrimination by adenylosuccinate synthetases.
Biochemistry, 45, 2006
|
|
1H7A
 
 | | Structural basis for allosteric substrate specificity regulation in class III ribonucleotide reductases: NRDD in complex with dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, ANAEROBIC RIBONUCLEOTIDE-TRIPHOSPHATE REDUCTASE LARGE CHAIN, FE (II) ION, ... | | Authors: | Larsson, K.-M, Andersson, J, Sjoeberg, B.-M, Nordlund, P, Logan, D.T. | | Deposit date: | 2001-07-04 | | Release date: | 2002-03-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis for Allosteric Substrate Specificty Regulation in Anaerobic Ribonucleotide Reductase
Structure, 9, 2001
|
|
1GX5
 
 | |
