7NNP
 
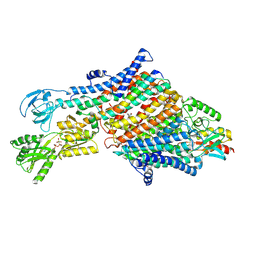 | | Rb-loaded cryo-EM structure of the E1-ATP KdpFABC complex. | | Descriptor: | CARDIOLIPIN, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Silberberg, J.M, Corey, R.A, Hielkema, L, Stock, C, Stansfeld, P.J, Paulino, C, Haenelt, I. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Deciphering ion transport and ATPase coupling in the intersubunit tunnel of KdpFABC.
Nat Commun, 12, 2021
|
|
6M2W
 
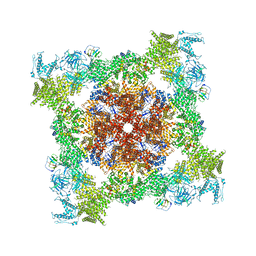 | | Structure of RyR1 (Ca2+/Caffeine/ATP/CaM1234/CHL) | | Descriptor: | 5-bromanyl-N-[4-chloranyl-2-methyl-6-(methylcarbamoyl)phenyl]-2-(3-chloranylpyridin-2-yl)pyrazole-3-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, ... | | Authors: | Ma, R, Haji-Ghassemi, O, Ma, D, Lin, L, Samurkas, A, Van Petegem, F, Yuchi, Z. | | Deposit date: | 2020-03-01 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for diamide modulation of ryanodine receptor.
Nat.Chem.Biol., 16, 2020
|
|
5IUJ
 
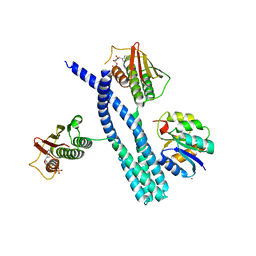 | | Crystal structure of the DesK-DesR complex in the phosphotransfer state with low Mg2+ (20 mM) | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Trajtenberg, F, Imelio, J.A, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2016-03-18 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Regulation of signaling directionality revealed by 3D snapshots of a kinase:regulator complex in action.
Elife, 5, 2016
|
|
7W6S
 
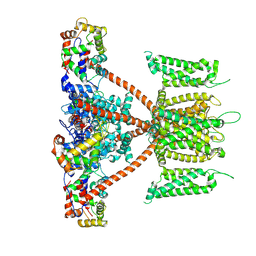 | | CryoEM structure of human KChIP2-Kv4.3 complex | | Descriptor: | Isoform 2 of Potassium voltage-gated channel subfamily D member 3, Kv channel-interacting protein 2 | | Authors: | Ma, D.M, Guo, J.T. | | Deposit date: | 2021-12-02 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for the gating modulation of Kv4.3 by auxiliary subunits.
Cell Res., 32, 2022
|
|
4XE5
 
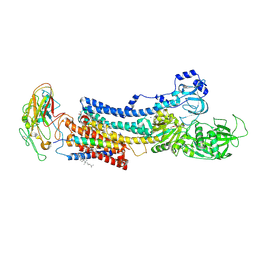 | | Crystal structure of the Na,K-ATPase from bovine | | Descriptor: | CHOLESTEROL, MAGNESIUM ION, OUABAIN, ... | | Authors: | Gregersen, J.L, Mattle, D, Fedosova, N.U, Nissen, P, Reinhard, L. | | Deposit date: | 2014-12-22 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.901 Å) | | Cite: | Isolation, crystallization and crystal structure determination of bovine kidney Na(+),K(+)-ATPase.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
7W6T
 
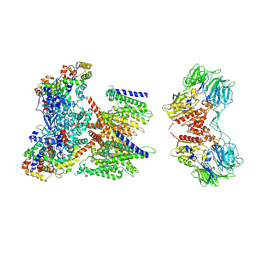 | | CryoEM structure of human KChIP1-Kv4.3-DPP6 complex | | Descriptor: | Dipeptidyl aminopeptidase-like protein 6, Isoform 2 of Potassium voltage-gated channel subfamily D member 3, Kv channel-interacting protein 1 | | Authors: | Ma, D.M, Guo, J.T. | | Deposit date: | 2021-12-02 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural basis for the gating modulation of Kv4.3 by auxiliary subunits.
Cell Res., 32, 2022
|
|
7W6N
 
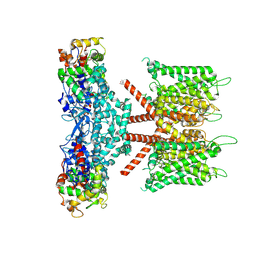 | | CryoEM structure of human KChIP1-Kv4.3 complex | | Descriptor: | Isoform 2 of Potassium voltage-gated channel subfamily D member 3, Kv channel-interacting protein 1 | | Authors: | Ma, D.M, Guo, J.T. | | Deposit date: | 2021-12-02 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for the gating modulation of Kv4.3 by auxiliary subunits.
Cell Res., 32, 2022
|
|
3ZQA
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF BETAINE ALDEHYDE DEHYDROGENASE MUTANT C286A FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH NADPH | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, BETAINE ALDEHYDE DEHYDROGENASE, ... | | Authors: | Diaz-Sanchez, A.G, Gonzalez-Segura, L, Rudino-Pinera, E, Lira-Rocha, A, Torres-Larios, A, Munoz-Clares, R.A. | | Deposit date: | 2011-06-08 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Novel Nadph-Cysteine Covalent Adduct Found in the Active Site of an Aldehyde Dehydrogenase.
Biochem.J., 439, 2011
|
|
1KQE
 
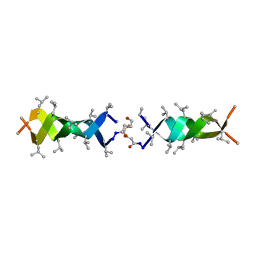 | | Solution structure of a linked shortened gramicidin A in benzene/acetone 10:1 | | Descriptor: | MINI-GRAMICIDIN A | | Authors: | Arndt, H.D, Bockelmann, D, Knoll, A, Lamberth, S, Griesinger, C, Koert, U. | | Deposit date: | 2002-01-05 | | Release date: | 2002-11-27 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Cation Control in Functional Helical Programming: Structures of a D,L-Peptide Ion Channel
Angew.Chem.Int.Ed.Engl., 41, 2002
|
|
6XA1
 
 | |
4WO2
 
 | |
5AVY
 
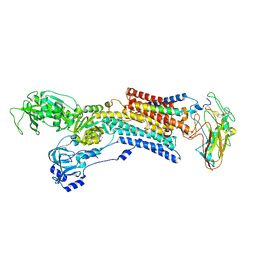 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 20 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
5AW0
 
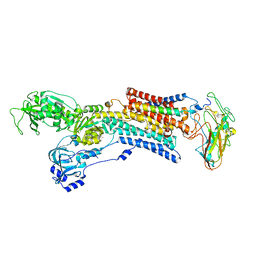 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 55 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
5AW6
 
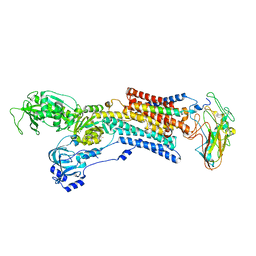 | | Kinetics by X-ray crystallography: Rb+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 5.5 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
8BFG
 
 | | Solution structure of human apo/Calmodulin G113R (G114R) | | Descriptor: | Calmodulin-1 | | Authors: | Wimmer, R, Holler, C.V, Petersson, N.M, Brohus, M.B, Niemelae, M, Overgaard, M.T, Iwai, H. | | Deposit date: | 2022-10-25 | | Release date: | 2023-10-04 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | Allosteric changes in protein stability and dynamics as pathogenic mechanism for calmodulin variants not affecting Ca 2+ coordinating residues.
Cell Calcium, 117, 2023
|
|
7W3Y
 
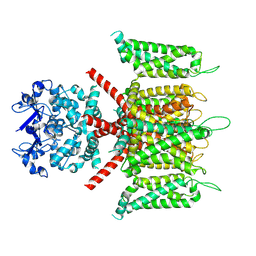 | | CryoEM structure of human Kv4.3 | | Descriptor: | Isoform 2 of Potassium voltage-gated channel subfamily D member 3 | | Authors: | Ma, D.M, Guo, J.T. | | Deposit date: | 2021-11-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the gating modulation of Kv4.3 by auxiliary subunits.
Cell Res., 32, 2022
|
|
7VVH
 
 | |
7VVD
 
 | |
4CSK
 
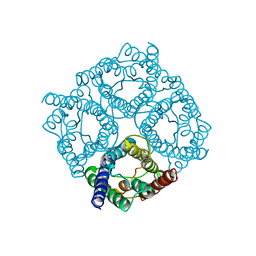 | | human Aquaporin | | Descriptor: | AQUAPORIN-1 | | Authors: | Ruiz-Carrillo, D, To-Yiu-Ying, J, Darwis, D, Soon, C.H, Cornvik, T, Torres, J, Lescar, J. | | Deposit date: | 2014-03-08 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Crystallization and Preliminary Crystallographic Analysis of Human Aquaporin 1 at a Resolution of 3.28 A.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
8BD2
 
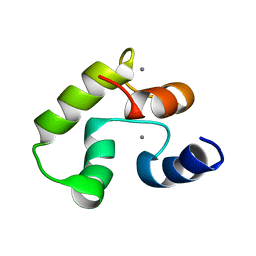 | | Calcium-bound Calmodulin variant G113R | | Descriptor: | CALCIUM ION, Calmodulin-3 | | Authors: | Wimmer, R, Holler, C.V, Petersson, N.M, Iwai, H, Niemelae, M.A, Brohus, M, Overgaard, M.T. | | Deposit date: | 2022-10-18 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | Allosteric changes in protein stability and dynamics as pathogenic mechanism for calmodulin variants not affecting Ca 2+ coordinating residues.
Cell Calcium, 117, 2023
|
|
1FQY
 
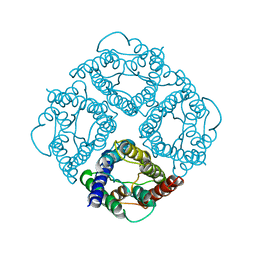 | | STRUCTURE OF AQUAPORIN-1 AT 3.8 A RESOLUTION BY ELECTRON CRYSTALLOGRAPHY | | Descriptor: | AQUAPORIN-1 | | Authors: | Murata, K, Mitsuoka, K, Hirai, T, Walz, T, Agre, P, Heymann, J.B, Engel, A, Fujiyoshi, Y. | | Deposit date: | 2000-09-07 | | Release date: | 2000-10-18 | | Last modified: | 2024-04-17 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.8 Å) | | Cite: | Structural determinants of water permeation through aquaporin-1.
Nature, 407, 2000
|
|
1Y62
 
 | | A 2.4 crystal structure of conkunitzin-S1, a novel Kunitz-fold cone snail neurotoxin. | | Descriptor: | Conkunitzin-S1, SULFATE ION | | Authors: | Dy, C.Y, Buczek, P, Horvath, M.P. | | Deposit date: | 2004-12-03 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of conkunitzin-S1, a neurotoxin and Kunitz-fold disulfide variant from cone snail.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
6Y94
 
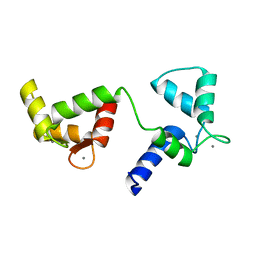 | | Ca2+-bound Calmodulin mutant N53I | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Holt, C, Nielsen, L.H, Lau, K, Brohus, M, Sorensen, A.B, Larsen, K.T, Sommer, C, Petegem, F.V, Overgaard, M.T, Wimmer, R. | | Deposit date: | 2020-03-06 | | Release date: | 2020-04-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The arrhythmogenic N53I variant subtly changes the structure and dynamics in the calmodulin N-terminal domain, altering its interaction with the cardiac ryanodine receptor.
J.Biol.Chem., 295, 2020
|
|
6Y95
 
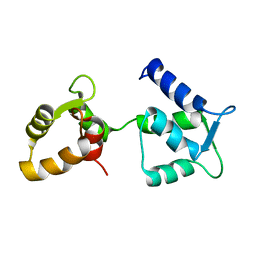 | | Ca2+-free Calmodulin mutant N53I | | Descriptor: | Calmodulin | | Authors: | Holt, C, Hamborg, L.N, Lau, K, Brohus, M, Sorensen, A.B, Larsen, K.T, Sommer, C, Petegem, F.V, Overgaard, M.T, Wimmer, R. | | Deposit date: | 2020-03-06 | | Release date: | 2020-04-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The arrhythmogenic N53I variant subtly changes the structure and dynamics in the calmodulin N-terminal domain, altering its interaction with the cardiac ryanodine receptor.
J.Biol.Chem., 295, 2020
|
|
6S5C
 
 | |
