4IAJ
 
 | |
1PPW
 
 | |
1IAM
 
 | | STRUCTURE OF THE TWO AMINO-TERMINAL DOMAINS OF HUMAN INTERCELLULAR ADHESION MOLECULE-1, ICAM-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, INTERCELLULAR ADHESION MOLECULE-1 | | Authors: | Bella, J, Kolatkar, P.R, Marlor, C, Greve, J.M, Rossmann, M.G. | | Deposit date: | 1998-02-22 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of the two amino-terminal domains of human ICAM-1 suggests how it functions as a rhinovirus receptor and as an LFA-1 integrin ligand.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
2NBM
 
 | |
6QGA
 
 | | Crystal structure of Ideonella sakaiensis MHETase bound to the non-hydrolyzable ligand MHETA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-hydroxyethylcarbamoyl)benzoic acid, CALCIUM ION, ... | | Authors: | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
6QGC
 
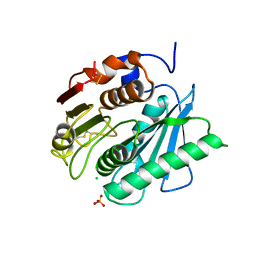 | | PETase from Ideonella sakaiensis without ligand | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
1PNZ
 
 | |
7XGA
 
 | |
1JOL
 
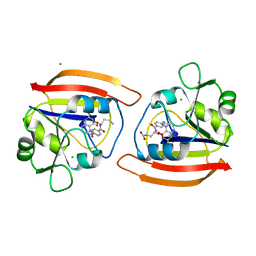 | | THE CRYSTAL STRUCTURE OF THE BINARY COMPLEX BETWEEN FOLINIC ACID (LEUCOVORIN) AND E. COLI DIHYDROFOLATE REDUCTASE | | Descriptor: | CALCIUM ION, CHLORIDE ION, DIHYDROFOLATE REDUCTASE, ... | | Authors: | Lee, H, Reyes, V.M, Kraut, J. | | Deposit date: | 1996-02-25 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structures of Escherichia coli dihydrofolate reductase complexed with 5-formyltetrahydrofolate (folinic acid) in two space groups: evidence for enolization of pteridine O4.
Biochemistry, 35, 1996
|
|
1JOM
 
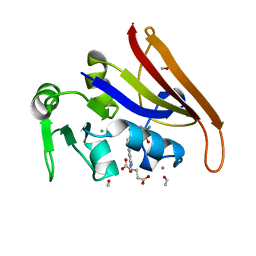 | | THE CRYSTAL STRUCTURE OF THE BINARY COMPLEX BETWEEN FOLINIC ACID (LEUCOVORIN) AND E. COLI DIHYDROFOLATE REDUCTASE | | Descriptor: | CALCIUM ION, CHLORIDE ION, DIHYDROFOLATE REDUCTASE, ... | | Authors: | Reyes, V.M, Lee, H, Kraut, J. | | Deposit date: | 1996-02-25 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Escherichia coli dihydrofolate reductase complexed with 5-formyltetrahydrofolate (folinic acid) in two space groups: evidence for enolization of pteridine O4.
Biochemistry, 35, 1996
|
|
2O7T
 
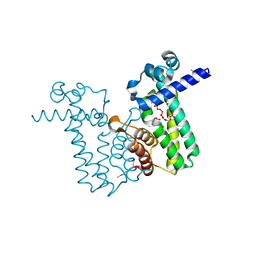 | |
5AVF
 
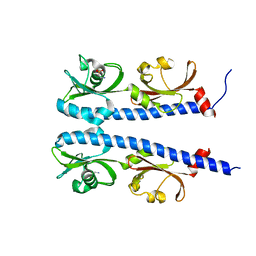 | | The ligand binding domain of Mlp37 with taurine | | Descriptor: | 2-AMINOETHANESULFONIC ACID, Methyl-accepting chemotaxis (MCP) signaling domain protein | | Authors: | Takahashi, Y, Sumita, K, Uchida, Y, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of a Vibrio cholerae chemoreceptor that senses taurine and amino acids as attractants
Sci Rep, 6, 2016
|
|
8QHP
 
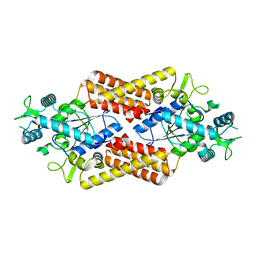 | |
2FTZ
 
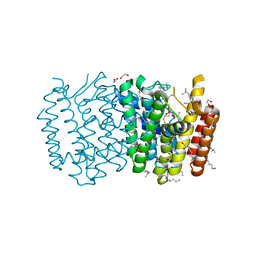 | |
4IAB
 
 | |
1T68
 
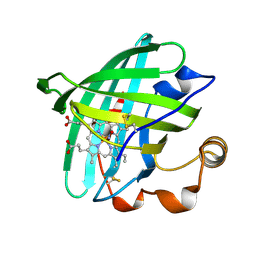 | | Crystal structure of nitrophorin 2 complex with NO | | Descriptor: | NITRIC OXIDE, Nitrophorin 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Weichsel, A, Montfort, W.R. | | Deposit date: | 2004-05-05 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Conformational change and heme ruffling in nitrophorin 2, a nitric oxide carrier from kissing bug
To be Published
|
|
3JR1
 
 | |
2A1O
 
 | |
3WMV
 
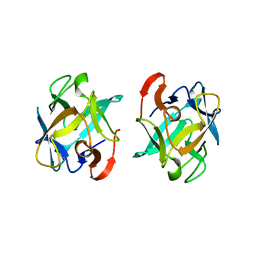 | | The structure of an anti-cancer lectin mytilec with ligand from the mussel Mytilus galloprovincialis | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Lectin | | Authors: | Terada, D, Kawai, F, Noguchi, H, Unzai, S, Park, S.-Y, Ozeki, Y, Tame, J.R.H. | | Deposit date: | 2013-11-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal structure of MytiLec, a galactose-binding lectin from the mussel Mytilus galloprovincialis with cytotoxicity against certain cancer cell types
Sci Rep, 6, 2016
|
|
4N90
 
 | |
3ATR
 
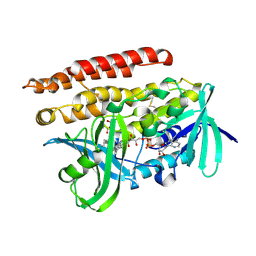 | | Geranylgeranyl Reductase (GGR) from Sulfolobus acidocaldarius co-crystallized with its ligand | | Descriptor: | Conserved Archaeal protein, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, PYROPHOSPHATE, ... | | Authors: | Fujihashi, M, Sasaki, D, Iwata, Y, Yoshimura, T, Hemmi, H, Miki, K. | | Deposit date: | 2011-01-12 | | Release date: | 2011-05-04 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mutation analysis of archaeal geranylgeranyl reductase
J.Mol.Biol., 409, 2011
|
|
3ENI
 
 | | Crystal structure of the Fenna-Matthews-Olson Protein from Chlorobaculum Tepidum | | Descriptor: | BACTERIOCHLOROPHYLL A, Bacteriochlorophyll a protein | | Authors: | Tronrud, D, Camara-Artigas, A, Blankenship, R, Allen, J.P. | | Deposit date: | 2008-09-25 | | Release date: | 2009-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structural basis for the difference in absorbance spectra for the FMO antenna protein from various green sulfur bacteria.
Photosynth.Res., 100, 2009
|
|
2ZEQ
 
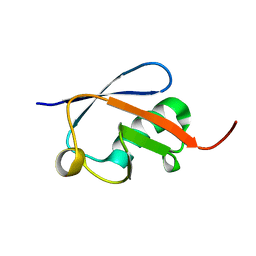 | | Crystal structure of ubiquitin-like domain of murine Parkin | | Descriptor: | E3 ubiquitin-protein ligase parkin | | Authors: | Tomoo, K. | | Deposit date: | 2007-12-14 | | Release date: | 2008-08-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure and molecular dynamics simulation of ubiquitin-like domain of murine parkin
Biochim.Biophys.Acta, 1784, 2008
|
|
2JIR
 
 | | A New Catalytic Mechanism of Periplasmic Nitrate Reductase from Desulfovibrio desulfuricans ATCC 27774 from Crystallographic and EPR Data and based on detailed analysis of the sixth ligand | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CYANIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Najmudin, S, Gonzalez, P.J, Trincao, J, Coelho, C, Mukhopadhyay, A, Romao, C.C, Moura, I, Moura, J.J.G, Brondino, C.D, Romao, M.J. | | Deposit date: | 2007-06-28 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Periplasmic Nitrate Reductase Revisited: A Sulfur Atom Completes the Sixth Coordination of the Catalytic Molybdenum.
J.Biol.Inorg.Chem., 13, 2008
|
|
2OUW
 
 | |
