5OTW
 
 | |
3C4O
 
 | | Crystal Structure of the SHV-1 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) E73M/S130K/S146M complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein, SULFATE ION | | Authors: | Reynolds, K.A, Hanes, M.S, Thomson, J.M, Antczak, A.J, Berger, J.M, Bonomo, R.A, Kirsch, J.F, Handel, T.M. | | Deposit date: | 2008-01-30 | | Release date: | 2008-05-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational redesign of the SHV-1 beta-lactamase/beta-lactamase inhibitor protein interface.
J.Mol.Biol., 382, 2008
|
|
2ANR
 
 | | Crystal structure (II) of Nova-1 KH1/KH2 domain tandem with 25nt RNA hairpin | | Descriptor: | 5'-R(*CP*(5BU)P*CP*GP*CP*GP*GP*AP*UP*CP*AP*GP*UP*CP*AP*CP*CP*CP*AP*AP*GP*CP*GP*AP*G)-3', MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Malinina, L, Teplova, M, Musunuru, K, Teplov, A, Darnell, J.C, Burley, S.K, Darnell, R.B, Patel, D.J. | | Deposit date: | 2005-08-11 | | Release date: | 2006-10-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Protein-RNA and protein-protein recognition by dual KH1/2 domains of the neuronal splicing factor Nova-1.
Structure, 19, 2011
|
|
3NOO
 
 | | Crystal Structure of C101A Isocyanide Hydratase from Pseudomonas fluorescens | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ThiJ/PfpI family protein | | Authors: | Lakshminarasimhan, M, Madzelan, P, Nan, R, Milkovic, N.M, Wilson, M.A. | | Deposit date: | 2010-06-25 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Evolution of New Enzymatic Function by Structural Modulation of Cysteine Reactivity in Pseudomonas fluorescens Isocyanide Hydratase.
J.Biol.Chem., 285, 2010
|
|
3BS9
 
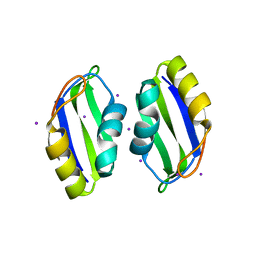 | | X-ray structure of human TIA-1 RRM2 | | Descriptor: | IODIDE ION, Nucleolysin TIA-1 isoform p40 | | Authors: | Kumar, A.O, Kielkopf, C.L. | | Deposit date: | 2007-12-22 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the central RNA recognition motif of human TIA-1 at 1.95A resolution.
Biochem.Biophys.Res.Commun., 367, 2008
|
|
1FG8
 
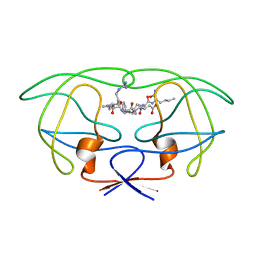 | | STRUCTURAL IMPLICATIONS OF DRUG RESISTANT MUTANTS OF HIV-1 PROTEASE: HIGH RESOLUTION CRYSTAL STRUCTURES OF THE MUTANT PROTEASE/SUBSTRATE ANALOG COMPLEXES | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-07-25 | | Release date: | 2001-06-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes.
Proteins, 43, 2001
|
|
1RRB
 
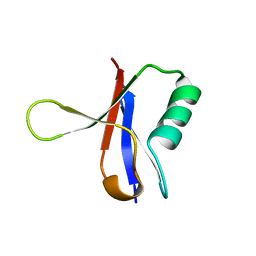 | | THE RAS-BINDING DOMAIN OF RAF-1 FROM RAT, NMR, 1 STRUCTURE | | Descriptor: | RAF PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE | | Authors: | Terada, T, Ito, Y, Shirouzu, M, Tateno, M, Hashimoto, K, Kigawa, T, Ebisuzaki, T, Takio, K, Shibata, T, Yokoyama, S, Smith, B.O, Laue, E.D, Cooper, J.A, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-03-26 | | Release date: | 1999-03-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance and molecular dynamics studies on the interactions of the Ras-binding domain of Raf-1 with wild-type and mutant Ras proteins.
J.Mol.Biol., 286, 1999
|
|
3DEA
 
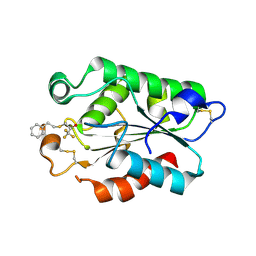 | | Glomerella cingulata PETFP-cutinase complex | | Descriptor: | 1,1,1-trifluoro-3-[(2-phenylethyl)sulfanyl]propan-2-one, Cutinase | | Authors: | Nyon, M.P, Rice, D.W, Berrisford, J.M, Hounslow, A.M, Moir, A.J.G, Huang, H, Nathan, S, Mahadi, N.M, Farah Diba, A.B, Craven, C.J. | | Deposit date: | 2008-06-09 | | Release date: | 2008-11-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalysis by Glomerella cingulata Cutinase Requires Conformational Cycling between the Active and Inactive States of Its Catalytic Triad
J.Mol.Biol., 385, 2009
|
|
2H0B
 
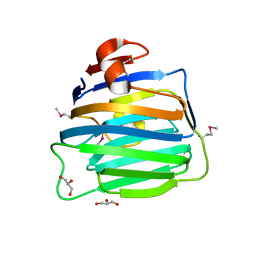 | | Crystal Structure of the second LNS/LG domain from Neurexin 1 alpha | | Descriptor: | CALCIUM ION, GLYCEROL, Neurexin-1-alpha | | Authors: | Sheckler, L.R, Henry, L, Sugita, S, Sudhof, T.C, Rudenko, G. | | Deposit date: | 2006-05-14 | | Release date: | 2006-06-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Second LNS/LG Domain from Neurexin 1{alpha}: Ca2+ binding and the effects of alternative splicing
J.Biol.Chem., 281, 2006
|
|
1RRG
 
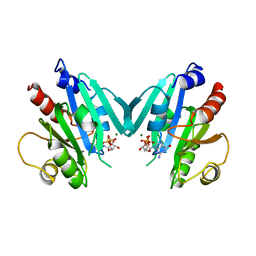 | | NON-MYRISTOYLATED RAT ADP-RIBOSYLATION FACTOR-1 COMPLEXED WITH GDP, DIMERIC CRYSTAL FORM | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAT ADP-RIBOSYLATION FACTOR-1 | | Authors: | Greasley, S.E, Jhoti, H, Bax, B. | | Deposit date: | 1995-12-16 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of rat ADP-ribosylation factor-1 (ARF-1) complexed to GDP determined from two different crystal forms.
Nat.Struct.Biol., 2, 1995
|
|
3DIW
 
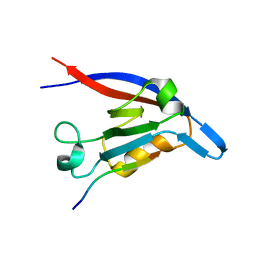 | | c-terminal beta-catenin bound TIP-1 structure | | Descriptor: | Tax1-binding protein 3, decameric peptide form Catenin beta-1 | | Authors: | Shen, Y. | | Deposit date: | 2008-06-21 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of beta-Catenin Recognition by Tax-interacting Protein-1
J.Mol.Biol., 384, 2008
|
|
3ZHX
 
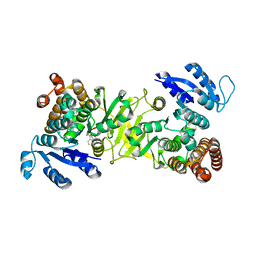 | | Structure of Mycobacterium tuberculosis DXR in complex with a fosmidomycin analogue | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, MANGANESE (II) ION, [(1S)-1-(3,4-dichlorophenyl)-3-[oxidanyl(phenylcarbonyl)amino]propyl]phosphonic acid | | Authors: | Bjorkelid, C, Jansson, A.M, Bergfors, T, Unge, T, Mowbray, S.L, Jones, T.A. | | Deposit date: | 2012-12-30 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dxr Inhibition by Potent Mono- and Disubstituted Fosmidomycin Analogues.
J.Med.Chem., 56, 2013
|
|
1BW5
 
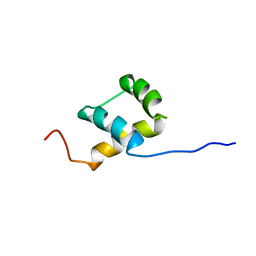 | | THE NMR SOLUTION STRUCTURE OF THE HOMEODOMAIN OF THE RAT INSULIN GENE ENHANCER PROTEIN ISL-1, 50 STRUCTURES | | Descriptor: | INSULIN GENE ENHANCER PROTEIN ISL-1 | | Authors: | Ippel, J.H, Larsson, G, Behravan, G, Zdunek, J, Lundqvist, M, Schleucher, J, Lycksell, P.-O, Wijmenga, S.S. | | Deposit date: | 1998-09-29 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the homeodomain of the rat insulin-gene enhancer protein isl-1. Comparison with other homeodomains.
J.Mol.Biol., 288, 1999
|
|
1RWV
 
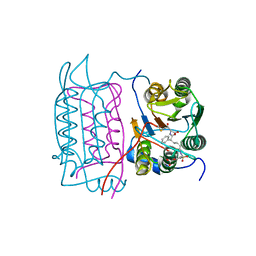 | |
1JQ9
 
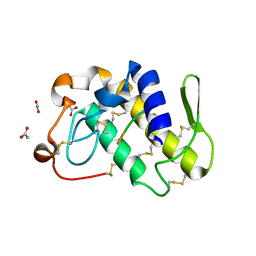 | | Crystal structure of a complex formed between phospholipase A2 from Daboia russelli pulchella and a designed pentapeptide Phe-Leu-Ser-Tyr-Lys at 1.8 resolution | | Descriptor: | ACETIC ACID, Peptide inhibitor, Phospholipase A2 | | Authors: | Chandra, V, Jasti, J, Kaur, P, Dey, S, Betzel, C, Singh, T.P. | | Deposit date: | 2001-08-04 | | Release date: | 2002-11-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Complex Formed between a Snake Venom Phospholipase A2 and a Potent Peptide Inhibitor Phe-Leu-Ser-Tyr-Lys at 1.8 A Resolution
J.BIOL.CHEM., 277, 2002
|
|
3SIX
 
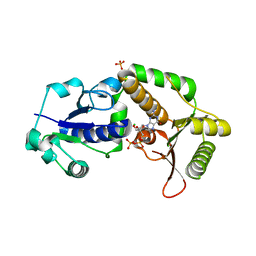 | | Crystal structure of NodZ alpha-1,6-fucosyltransferase soaked with GDP-fucose | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, Nodulation fucosyltransferase NodZ, ... | | Authors: | Brzezinski, K, Dauter, Z, Jaskolski, M. | | Deposit date: | 2011-06-20 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of NodZ alpha-1,6-fucosyltransferase in complex with GDP and GDP-fucose
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3SMH
 
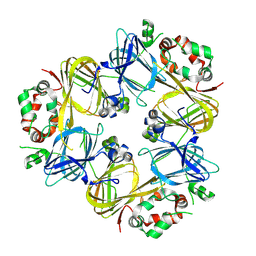 | |
2OQB
 
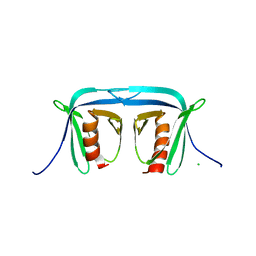 | |
4FWW
 
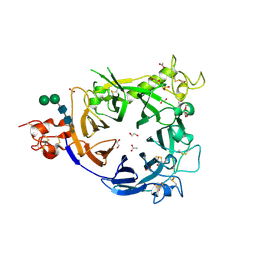 | |
3TX7
 
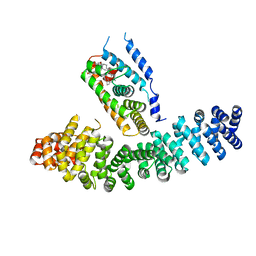 | | Crystal structure of LRH-1/beta-catenin complex | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, Catenin beta-1, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Yumoto, F, Fletterick, R. | | Deposit date: | 2011-09-22 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural basis of coactivation of liver receptor homolog-1 by beta-catenin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3N0Q
 
 | |
4OEM
 
 | | Crystal structure of Cathepsin C in complex with dipeptide substrates | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zhao, B, Smallwood, A, Concha, N. | | Deposit date: | 2014-01-13 | | Release date: | 2015-03-25 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The amino-acid substituents of dipeptide substrates of cathepsin C can determine the rate-limiting steps of catalysis.
Biochemistry, 51, 2012
|
|
3N45
 
 | | Human FPPS complex with FBS_04 and zoledronic acid/MG2+ | | Descriptor: | (2S)-1-[(benzyloxy)carbonyl]-2,3-dihydro-1H-indole-2-carboxylic acid, FARNESYL PYROPHOSPHATE SYNTHASE, MAGNESIUM ION, ... | | Authors: | Rondeau, J.-M. | | Deposit date: | 2010-05-21 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Allosteric non-bisphosphonate FPPS inhibitors identified by fragment-based discovery.
Nat.Chem.Biol., 6, 2010
|
|
1ZA4
 
 | |
1FFI
 
 | | STRUCTURAL IMPLICATIONS OF DRUG RESISTANT MUTANTS OF HIV-1 PROTEASE: HIGH RESOLUTION CRYSTAL STRUCTURES OF THE MUTANT PROTEASE/SUBSTRATE ANALOG COMPLEXES | | Descriptor: | N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-07-25 | | Release date: | 2001-06-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes.
Proteins, 43, 2001
|
|
