3S9Q
 
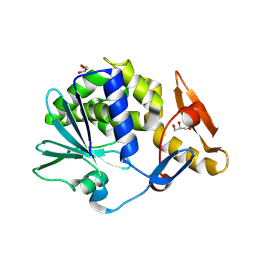 | | Crystal structure of native type 1 ribosome inactivating protein from Momordica balsamina at 1.67 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein | | Authors: | Pandey, N, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-06-02 | | Release date: | 2011-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structures of a type-1 ribosome inactivating protein from Momordica balsamina in the bound and unbound states.
Biochim.Biophys.Acta, 1824, 2012
|
|
1EXQ
 
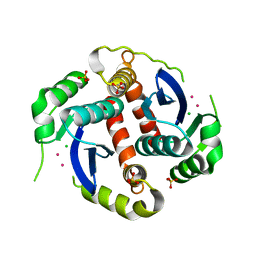 | | CRYSTAL STRUCTURE OF THE HIV-1 INTEGRASE CATALYTIC CORE DOMAIN | | Descriptor: | CADMIUM ION, CHLORIDE ION, POL POLYPROTEIN, ... | | Authors: | Chen, J.C.-H, Krucinski, J, Miercke, L.J.W, Finer-Moore, J.S, Tang, A.H, Leavitt, A.D, Stroud, R.M. | | Deposit date: | 2000-05-03 | | Release date: | 2000-11-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
3SG1
 
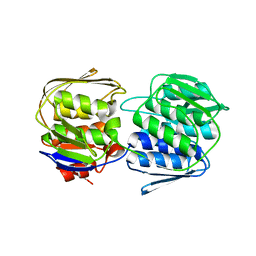 | | 2.6 Angstrom Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase 1 (MurA1) from Bacillus anthracis | | Descriptor: | TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, UDP-N-acetylglucosamine 1-carboxyvinyltransferase 1 | | Authors: | Minasov, G, Halavaty, A, Filippova, E.V, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-14 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.6 Angstrom Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase 1 (MurA1) from Bacillus anthracis.
TO BE PUBLISHED
|
|
3R05
 
 | |
4H45
 
 | |
2HAK
 
 | | Catalytic and ubiqutin-associated domains of MARK1/PAR-1 | | Descriptor: | Serine/threonine-protein kinase MARK1 | | Authors: | Marx, A, Nugoor, C, Mueller, J, Panneerselvam, S, Mandelkow, E.-M, Mandelkow, E. | | Deposit date: | 2006-06-13 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural variations in the catalytic and ubiquitin-associated domains of microtubule-associated protein/microtubule affinity regulating kinase (MARK) 1 and MARK2
J.Biol.Chem., 281, 2006
|
|
2EWM
 
 | |
3KDD
 
 | | Crystal Structure of HIV-1 Protease (Q7K, L33I, L63I) in Complex with KNI-10265 | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(2,6-difluorophenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H- inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, Protease | | Authors: | Chufan, E.E, Kawasaki, Y, Freire, E, Amzel, L.M. | | Deposit date: | 2009-10-22 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | How much binding affinity can be gained by filling a cavity?
Chem.Biol.Drug Des., 75, 2010
|
|
3WIZ
 
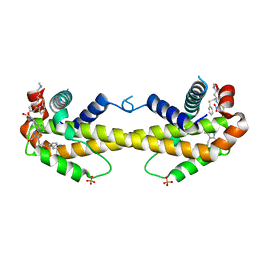 | | Crystal structure of Bcl-xL in complex with compound 10 | | Descriptor: | 7-(4-{[(4-{[(2R)-4-(dimethylamino)-1-(phenylsulfanyl)butan-2-yl]amino}-3-nitrophenyl)sulfonyl]carbamoyl}-2-methylphenyl)-3-[3-(naphthalen-1-yloxy)propyl]pyrazolo[1,5-a]pyridine-2-carboxylic acid, Bcl-2-like protein 1, PHOSPHATE ION | | Authors: | Sogabe, S, Igaki, S, Hayano, Y. | | Deposit date: | 2013-09-26 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Discovery of potent Mcl-1/Bcl-xL dual inhibitors by using a hybridization strategy based on structural analysis of target proteins.
J.Med.Chem., 56, 2013
|
|
2KTV
 
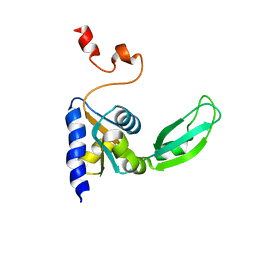 | |
2KNT
 
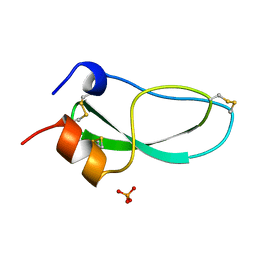 | |
1ZCL
 
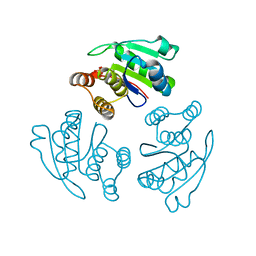 | | prl-1 c104s mutant in complex with sulfate | | Descriptor: | SULFATE ION, protein tyrosine phosphatase 4a1 | | Authors: | Sun, J.P, Wang, W.Q, Yang, H, Liu, S, Liang, F, Fedorov, A.A, Almo, S.C, Zhang, Z.Y. | | Deposit date: | 2005-04-12 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and Biochemical Properties of PRL-1, a Phosphatase Implicated in Cell Growth, Differentiation, and Tumor Invasion.
Biochemistry, 44, 2005
|
|
1JP4
 
 | | Crystal Structure of an Enzyme Displaying both Inositol-Polyphosphate 1-Phosphatase and 3'-Phosphoadenosine-5'-Phosphate Phosphatase Activities | | Descriptor: | 3'(2'),5'-bisphosphate nucleotidase, ADENOSINE MONOPHOSPHATE, BETA-MERCAPTOETHANOL, ... | | Authors: | Patel, S, Yenush, L, Rodriguez, P.L, Serrano, R, Blundell, T.L. | | Deposit date: | 2001-08-01 | | Release date: | 2001-08-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of an enzyme displaying both inositol-polyphosphate-1-phosphatase and 3'-phosphoadenosine-5'-phosphate phosphatase activities: a novel target of lithium therapy.
J.Mol.Biol., 315, 2002
|
|
3O56
 
 | | Catalytic domain of human phosphodiesterase 4b2b in complex with a 5-heterocycle pyrazolopyridine inhibitor | | Descriptor: | 1-ethyl-5-[3-(2-oxo-2-pyrrolidin-1-ylethyl)-1,2,4-oxadiazol-5-yl]-N-(tetrahydro-2H-pyran-4-yl)-1H-pyrazolo[3,4-b]pyridin-4-amine, ARSENIC, GLYCEROL, ... | | Authors: | Somers, D.O, Neu, M. | | Deposit date: | 2010-07-28 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Pyrazolopyridines as potent PDE4B inhibitors: 5-heterocycle SAR.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1AGM
 
 | | Refined structure for the complex of acarbose with glucoamylase from Aspergillus awamori var. x100 to 2.4 angstroms resolution | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLUCOAMYLASE-471, alpha-D-mannopyranose, ... | | Authors: | Aleshin, A.E, Firsov, L.M, Honzatko, R.B. | | Deposit date: | 1994-05-13 | | Release date: | 1994-09-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined structure for the complex of acarbose with glucoamylase from Aspergillus awamori var. X100 to 2.4-A resolution.
J.Biol.Chem., 269, 1994
|
|
1WQ8
 
 | | Crystal structure of Vammin, a VEGF-F from a snake venom | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Vascular endothelial growth factor toxin | | Authors: | Suto, K, Yamazaki, Y, Morita, T, Mizuno, H. | | Deposit date: | 2004-09-23 | | Release date: | 2004-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of novel vascular endothelial growth factors (VEGF) from snake venoms: insight into selective VEGF binding to kinase insert domain-containing receptor but not to fms-like tyrosine kinase-1.
J.Biol.Chem., 280, 2005
|
|
3SLS
 
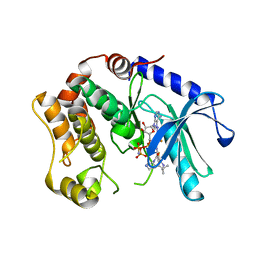 | | Crystal Structure of human MEK-1 kinase in complex with UCB1353770 and AMPPNP | | Descriptor: | 2-[(2-fluoro-4-iodophenyl)amino]-5,5-dimethyl-8-oxo-N-[(3R)-piperidin-3-yl]-5,6,7,8-tetrahydro-4H-thieno[2,3-c]azepine-3-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Meier, C, Ceska, T.A. | | Deposit date: | 2011-06-25 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Engineering human MEK-1 for structural studies: A case study of combinatorial domain hunting.
J.Struct.Biol., 177, 2012
|
|
1RRI
 
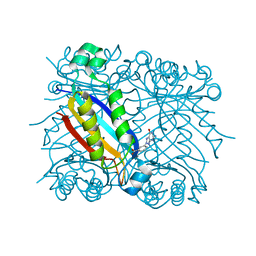 | | DHNA complex with 3-(5-amino-7-hydroxy-[1,2,3] triazolo [4,5-d]pyrimidin-2-yl)-benzoic acid | | Descriptor: | 3-(5-AMINO-7-HYDROXY-[1,2,3]TRIAZOLO[4,5-D]PYRIMIDIN-2-YL)-BENZOIC ACID, Dihydroneopterin aldolase | | Authors: | Sanders, W.J, Nienaber, V.L, Lerner, C.G, McCall, J.O, Merrick, S.M, Swanson, S.J, Harlan, J.E, Stoll, V.S, Stamper, G.F, Betz, S.F, Condroski, K.R, Meadows, R.P, Severin, J.M, Walter, K.A, Magdalinos, P, Jakob, C.G, Wagner, R, Beutel, B.A. | | Deposit date: | 2003-12-08 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Potent Inhibitors of Dihydroneopterin Aldolase Using CrystaLEAD High-Throughput X-ray Crystallographic Screening and Structure-Directed Lead Optimization.
J.Med.Chem., 47, 2004
|
|
1RWO
 
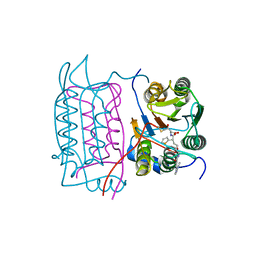 | | Crystal structure of human caspase-1 in complex with 4-oxo-3-{6-[4-(quinoxalin-2-ylamino)-benzoylamino]-2-thiophen-2-yl-hexanoylamino}-pentanoic acid | | Descriptor: | 4-OXO-3-{6-[4-(QUINOXALIN-2-YLAMINO)-BENZOYLAMINO]-2-THIOPHEN-2-YL-HEXANOYLAMINO}-PENTANOIC ACID, Interleukin-1 beta convertase | | Authors: | Romanowski, M.J, Waal, N.D, Fahr, B.T, O'Brien, T. | | Deposit date: | 2003-12-16 | | Release date: | 2004-12-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of caspase-1 inhibitors derived from Tethering.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1UR8
 
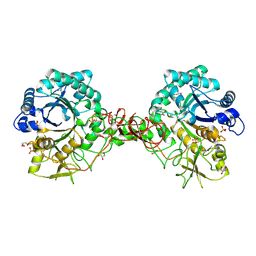 | | Interactions of a family 18 chitinase with the designed inhibitor HM508, and its degradation product, chitobiono-delta-lactone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-(acetylamido)-2-deoxy-D-glucono-1,5-lactone, CHITINASE B, GLYCEROL, ... | | Authors: | Vaaje-Kolstad, G, Vasella, A, Peter, M.G, Netter, C, Houston, D.R, Westereng, B, Synstad, B, Eijsink, V.G.H, Van Aalten, D.M.F. | | Deposit date: | 2003-10-27 | | Release date: | 2004-04-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interactions of a Family 18 Chitinase with the Designed Inhibitor Hm508 and its Degradation Product, Chitobiono-Delta-Lactone.
J.Biol.Chem., 279, 2004
|
|
1GFW
 
 | |
7LT9
 
 | |
2CFE
 
 | | The 1.5 A crystal structure of the Malassezia sympodialis Mala s 6 allergen, a member of the cyclophilin pan-allergen family | | Descriptor: | ALANINE, ALLERGEN, GLYCEROL, ... | | Authors: | Limacher, A, Glaser, A.G, Fluckiger, S, Scheynius, A, Scapozza, L, Crameri, R. | | Deposit date: | 2006-02-20 | | Release date: | 2006-02-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Analysis of the cross-reactivity and of the 1.5 A crystal structure of the Malassezia sympodialis Mala s 6 allergen, a member of the cyclophilin pan-allergen family.
Biochem. J., 396, 2006
|
|
3PFE
 
 | |
2AVW
 
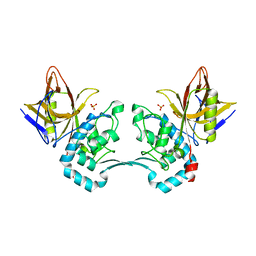 | | Crystal structure of monoclinic form of streptococcus Mac-1 | | Descriptor: | GLYCEROL, IgG-degrading protease, SULFATE ION | | Authors: | Agniswamy, J, Nagiec, M.J, Liu, M, Schuck, P, Musser, J.M, Sun, P.D. | | Deposit date: | 2005-08-30 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of group a streptococcus mac-1: insight into dimer-mediated specificity for recognition of human IgG.
Structure, 14, 2006
|
|
