1FUK
 
 | |
7UXZ
 
 | | Crystal structure of SARS-CoV-2 nucleocapsid protein C-terminal domain complexed with Chicoric acid | | Descriptor: | (2R,3R)-2,3-bis{[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy}butanedioic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bezerra, E.H.S, Tonoli, C.C.C, Soprano, A.S, Franchini, K.G, Trivella, D.B.B, Benedetti, C.E. | | Deposit date: | 2022-05-06 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | Discovery and structural characterization of chicoric acid as a SARS-CoV-2 nucleocapsid protein ligand and RNA binding disruptor.
Sci Rep, 12, 2022
|
|
2IDR
 
 | | Crystal structure of translation initiation factor EIF4E from wheat | | Descriptor: | Eukaryotic translation initiation factor 4E-1 | | Authors: | Monzingo, A.F, Sadow, J, Dhaliwal, S, Lyon, A, Hoffman, D.W, Robertus, J.D, Browning, K.S. | | Deposit date: | 2006-09-15 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of eukaryotic translation initiation factor-4E from wheat reveals a novel disulfide bond.
Plant Physiol., 143, 2007
|
|
5CXB
 
 | |
4Z2X
 
 | |
5CXC
 
 | | Structure of Ytm1 bound to the C-terminal domain of Erb1 in P 65 2 2 space group | | Descriptor: | CHLORIDE ION, Ribosome biogenesis protein ERB1, Ribosome biogenesis protein YTM1 | | Authors: | Wegrecki, M, Bravo, J. | | Deposit date: | 2015-07-28 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of Erb1-Ytm1 complex reveals the functional importance of a high-affinity binding between two beta-propellers during the assembly of large ribosomal subunits in eukaryotes.
Nucleic Acids Res., 43, 2015
|
|
5CYK
 
 | | Structure of Ytm1 bound to the C-terminal domain of Erb1-R486E | | Descriptor: | CHLORIDE ION, Ribosome biogenesis protein ERB1, Ribosome biogenesis protein YTM1 | | Authors: | Wegrecki, M, Bravo, J. | | Deposit date: | 2015-07-30 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of Erb1-Ytm1 complex reveals the functional importance of a high-affinity binding between two beta-propellers during the assembly of large ribosomal subunits in eukaryotes.
Nucleic Acids Res., 43, 2015
|
|
5T5A
 
 | |
4CZY
 
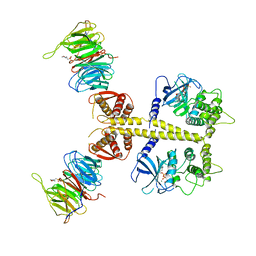 | | Complex of Neurospora crassa PAN2 (WD40-CS1) with PAN3 (pseudokinase and C-term) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, PAB-DEPENDENT POLY(A)-SPECIFIC RIBONUCLEASE SUBUNIT PAN2, ... | | Authors: | Jonas, S, Izaurralde, E, Weichenrieder, O. | | Deposit date: | 2014-04-22 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | An Asymmetric Pan3 Dimer Recruits a Single Pan2 Exonuclease to Mediate Mrna Deadenylation and Decay.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4D0K
 
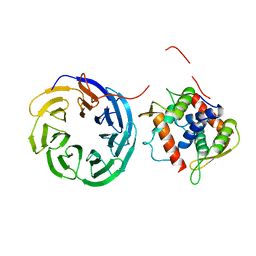 | |
6A7V
 
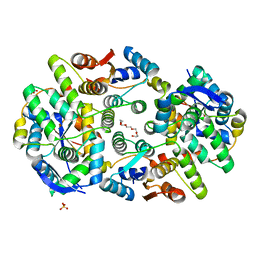 | | Crystal structure of Mycobacterium tuberculosis VapBC11 toxin-antitoxin complex | | Descriptor: | Antitoxin VapB11, PENTAETHYLENE GLYCOL, Ribonuclease VapC11, ... | | Authors: | Deep, A, Thakur, K.G. | | Deposit date: | 2018-07-04 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural, functional and biological insights into the role of Mycobacterium tuberculosis VapBC11 toxin-antitoxin system: targeting a tRNase to tackle mycobacterial adaptation.
Nucleic Acids Res., 46, 2018
|
|
1QD2
 
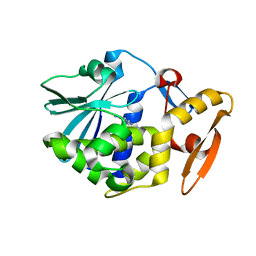 | |
4FJS
 
 | | Crystal structure of ureidoglycolate dehydrogenase enzyme in apo form | | Descriptor: | Ureidoglycolate dehydrogenase | | Authors: | Kim, M.I, Shin, I, Lee, J, Rhee, S. | | Deposit date: | 2012-06-12 | | Release date: | 2013-01-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural and functional insights into (s)-ureidoglycolate dehydrogenase, a metabolic branch point enzyme in nitrogen utilization.
Plos One, 7, 2012
|
|
4CZX
 
 | | Complex of Neurospora crassa PAN2 (WD40) with PAN3 (C-TERM) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, PAB-DEPENDENT POLY(A)-SPECIFIC RIBONUCLEASE SUBUNIT PAN2, ... | | Authors: | Jonas, S, Izaurralde, E, Weichenrieder, O. | | Deposit date: | 2014-04-22 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An Asymmetric Pan3 Dimer Recruits a Single Pan2 Exonuclease to Mediate Mrna Deadenylation and Decay.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4NMG
 
 | |
3UCG
 
 | |
1IG1
 
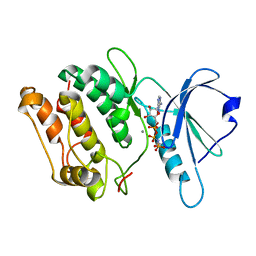 | | 1.8A X-Ray structure of ternary complex of a catalytic domain of death-associated protein kinase with ATP analogue and Mn. | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, death-associated protein kinase | | Authors: | Tereshko, V, Teplova, M, Brunzelle, J, Watterson, D.M, Egli, M. | | Deposit date: | 2001-04-16 | | Release date: | 2002-04-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the catalytic domain of human protein kinase associated with apoptosis and tumor suppression.
Nat.Struct.Biol., 8, 2001
|
|
4BSN
 
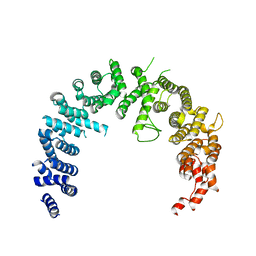 | | Crystal structure of the Nuclear Export Receptor CRM1 (exportin-1) lacking the C-terminal helical extension at 4.1A | | Descriptor: | EXPORTIN-1 | | Authors: | Dian, C, Bernaudat, F, Langer, K, Oliva, M.F, Fornerod, M, Schoehn, G, Muller, C.W, Petosa, C. | | Deposit date: | 2013-06-11 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structure of a Truncation Mutant of the Nuclear Export Factor Crm1 Provides Insights Into the Auto-Inhibitory Role of its C-Terminal Helix.
Structure, 21, 2013
|
|
7QB3
 
 | |
3IVN
 
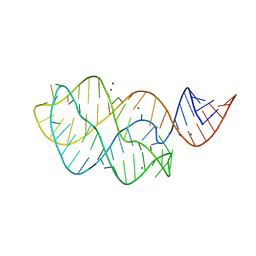 | | Structure of the U65C mutant A-riboswitch aptamer from the Bacillus subtilis pbuE operon | | Descriptor: | A-riboswitch, BROMIDE ION, MAGNESIUM ION | | Authors: | Delfosse, V, Dagenais, P, Chausse, D, Di Tomasso, G, Legault, P. | | Deposit date: | 2009-09-01 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Riboswitch structure: an internal residue mimicking the purine ligand.
Nucleic Acids Res., 38, 2010
|
|
1AH9
 
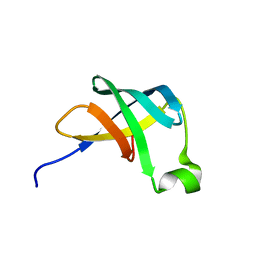 | | THE STRUCTURE OF THE TRANSLATIONAL INITIATION FACTOR IF1 FROM ESCHERICHIA COLI, NMR, 19 STRUCTURES | | Descriptor: | INITIATION FACTOR 1 | | Authors: | Sette, M, Van Tilborg, P, Spurio, R, Kaptein, R, Paci, M, Gualerzi, C.O, Boelens, R. | | Deposit date: | 1997-04-16 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the translational initiation factor IF1 from E.coli contains an oligomer-binding motif.
EMBO J., 16, 1997
|
|
1R4H
 
 | | NMR Solution structure of the IIIc domain of GB Virus B IRES Element | | Descriptor: | 5'-R(*GP*GP*GP*CP*AP*AP*GP*CP*CP*C)-3' | | Authors: | Kaluarachchi, K, Thiviyanathan, V, Rijinbrand, R, Lemon, S.M, Gorenstein, D.G. | | Deposit date: | 2003-10-06 | | Release date: | 2004-10-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mutational and structural analysis of stem-loop IIIC of the hepatitis C virus and GB virus B internal ribosome entry sites.
J.Mol.Biol., 343, 2004
|
|
7UXX
 
 | | Crystal structure of SARS-CoV-2 nucleocapsid protein C-terminal domain | | Descriptor: | ACETATE ION, GLYCEROL, Nucleoprotein | | Authors: | Bezerra, E.H.S, Tonoli, C.C.C, Soprano, A.S, Franchini, K.G, Trivella, D.B.B, Benedetti, C.E. | | Deposit date: | 2022-05-06 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and structural characterization of chicoric acid as a SARS-CoV-2 nucleocapsid protein ligand and RNA binding disruptor.
Sci Rep, 12, 2022
|
|
7AFR
 
 | | Ribosome maturation factor RimP (apo) | | Descriptor: | Ribosome maturation factor RimP | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Fucini, P, Connell, S. | | Deposit date: | 2020-09-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
2OML
 
 | | crystal structure of E. coli pseudouridine synthase RluE | | Descriptor: | Ribosomal large subunit pseudouridine synthase E, SULFATE ION | | Authors: | Pan, H, Ho, J.D, Stroud, R.M, Finer-Moore, J. | | Deposit date: | 2007-01-22 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Crystal Structure of E. coli rRNA Pseudouridine Synthase RluE.
J.Mol.Biol., 367, 2007
|
|
