8BLU
 
 | | The PDZ domains of human SDCBP | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ALANINE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Daniel-Mozo, M, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2022-11-10 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The PDZ domains of human SDCBP
To Be Published
|
|
1U2C
 
 | |
8AX5
 
 | | Crystal structure of a CGRP receptor ectodomain heterodimer bound to macrocyclic inhibitor HTL0029881 | | Descriptor: | (1~{R},10~{R},20~{E})-12-methyl-10-[(7-methyl-2~{H}-indazol-5-yl)methyl]-15,18-dioxa-9,12,24,26-tetrazapentacyclo[20.5.2.1^{1,4}.1^{3,7}.0^{25,28}]hentriaconta-3,5,7(30),20,22,24,28-heptaene-8,11,27-trione, Maltose/maltodextrin-binding periplasmic protein,Receptor activity-modifying protein 1,Calcitonin gene-related peptide type 1 receptor, TETRAETHYLENE GLYCOL, ... | | Authors: | Southall, S.M, Watson, S.P. | | Deposit date: | 2022-08-30 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Novel Macrocyclic Antagonists of the CGRP Receptor Part 2: Stereochemical Inversion Induces an Unprecedented Binding Mode.
Acs Med.Chem.Lett., 13, 2022
|
|
1UF5
 
 | | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase complexed with N-carbamyl-D-methionine | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYLSULFANYL-2-UREIDO-BUTYRIC ACID, N-carbamyl-D-amino acid amidohydrolase | | Authors: | Hashimoto, H, Aoki, M, Shimizu, T, Nakai, T, Morikawa, H, Ikenaka, Y, Takahashi, S, Sato, M. | | Deposit date: | 2003-05-23 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of C171A/V236A mutant of N-carbamyl-D-amino acid amidohydrolase
To be published
|
|
8B6Z
 
 | |
8B40
 
 | |
1UFA
 
 | | Crystal structure of TT1467 from Thermus thermophilus HB8 | | Descriptor: | TT1467 protein | | Authors: | Idaka, M, Terada, T, Murayama, K, Yamaguchi, H, Nureki, O, Ishitani, R, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-28 | | Release date: | 2003-11-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of TT1467 from Thermus thermophilus HB8
To be published
|
|
3S92
 
 | | Crystal Structure of the second bromodomain of human BRD3 in complex with the inhibitor JQ1 | | Descriptor: | (6S)-6-(2-tert-butoxy-2-oxoethyl)-4-(4-chlorophenyl)-2,3,9-trimethyl-6,7-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, 1,2-ETHANEDIOL, Bromodomain-containing protein 3 | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Keates, T, Felletar, I, Fedorov, O, Muniz, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Bradner, J.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-05-31 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal Structure of the second bromodomain of human BRD3 in complex with the inhibitor JQ1
To be Published
|
|
1UFO
 
 | | Crystal Structure of TT1662 from Thermus thermophilus | | Descriptor: | hypothetical protein TT1662 | | Authors: | Murayama, K, Shirouzu, M, Terada, T, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-03 | | Release date: | 2003-12-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of TT1662 from Thermus thermophilus HB8: a member of the alpha/beta hydrolase fold enzymes.
Proteins, 58, 2005
|
|
8BEN
 
 | | LRR domain Structure of the LRRC8C protein | | Descriptor: | Volume-regulated anion channel subunit LRRC8C | | Authors: | Sawicka, M, Dutzler, R. | | Deposit date: | 2022-10-21 | | Release date: | 2022-12-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of a volume-regulated heteromeric LRRC8A/C channel.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3S9L
 
 | | Complex between transferrin receptor 1 and transferrin with iron in the N-Lobe, cryocooled 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CARBONATE ION, ... | | Authors: | Eckenroth, B.E, Steere, A.N, Mason, A.B, Everse, S.J. | | Deposit date: | 2011-06-01 | | Release date: | 2011-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | How the binding of human transferrin primes the transferrin receptor potentiating iron release at endosomal pH.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8B41
 
 | |
1UGR
 
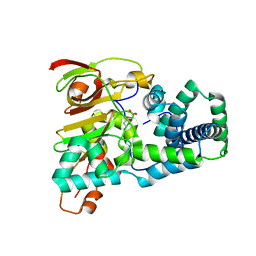 | | Crystal structure of aT109S mutant of Co-type nitrile hydratase | | Descriptor: | COBALT (II) ION, Nitrile Hydratase alpha subunit, Nitrile Hydratase beta subunit | | Authors: | Miyanaga, A, Fushinobu, S, Ito, K, Shoun, H, Wakagi, T. | | Deposit date: | 2003-06-17 | | Release date: | 2004-06-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mutational and structural analysis of cobalt-containing nitrile hydratase on substrate and metal binding
Eur.J.Biochem., 271, 2004
|
|
8AT6
 
 | | Cryo-EM structure of yeast Elp456 subcomplex | | Descriptor: | Elongator complex protein 4, Elongator complex protein 5, Elongator complex protein 6 | | Authors: | Jaciuk, M, Scherf, D, Kaszuba, K, Gaik, M, Koscielniak, A, Krutyholowa, R, Rawski, M, Indyka, P, Biela, A, Dobosz, D, Lin, T.-Y, Abbassi, N, Hammermeister, A, Chramiec-Glabik, A, Kosinski, J, Schaffrath, R, Glatt, S. | | Deposit date: | 2022-08-22 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the fully assembled Elongator complex.
Nucleic Acids Res., 51, 2023
|
|
1U2P
 
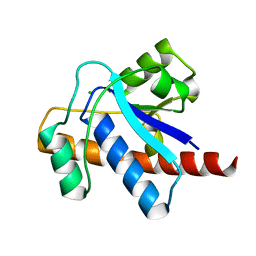 | | Crystal structure of Mycobacterium tuberculosis Low Molecular Protein Tyrosine Phosphatase (MPtpA) at 1.9A resolution | | Descriptor: | CHLORIDE ION, low molecular weight protein-tyrosine-phosphatase | | Authors: | Madhurantakam, C, Rajakumara, E, Mazumdar, P.A, Saha, B, Mitra, D, Wiker, H.G, Sankaranarayanan, R, Das, A.K. | | Deposit date: | 2004-07-20 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Low-Molecular-Weight Protein Tyrosine Phosphatase from Mycobacterium tuberculosis at 1.9-A Resolution
J.Bacteriol., 187, 2005
|
|
8B9F
 
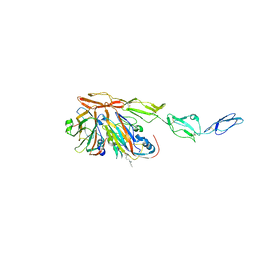 | | Structure of Echovirus 11 complexed with DAF (CD55) calculated from symmetry expansion | | Descriptor: | Complement decay-accelerating factor, Genome polyprotein, SPHINGOSINE | | Authors: | Stuart, D.I, Ren, J, Qin, L, Zhou, D. | | Deposit date: | 2022-10-05 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Switching of Receptor Binding Poses between Closely Related Enteroviruses.
Viruses, 14, 2022
|
|
3SB4
 
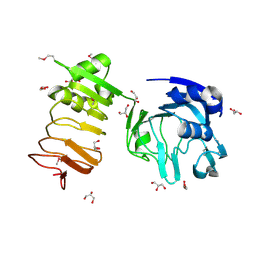 | |
1U2X
 
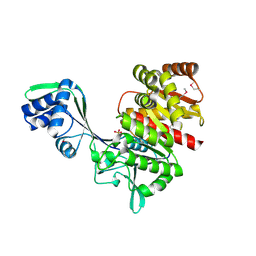 | | Crystal Structure of a Hypothetical ADP-dependent Phosphofructokinase from Pyrococcus horikoshii OT3 | | Descriptor: | ADP-specific phosphofructokinase, SULFATE ION | | Authors: | Wong, A.H.Y, Jia, Z, Skarina, T, Walker, J.R, Arrowsmith, C, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ADP-dependent 6-phosphofructokinase from Pyrococcus horikoshii OT3: structure determination and biochemical characterization of PH1645.
J.Biol.Chem., 284, 2009
|
|
1U0S
 
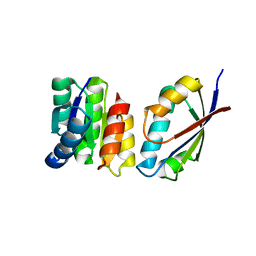 | | Chemotaxis kinase CheA P2 domain in complex with response regulator CheY from the thermophile thermotoga maritima | | Descriptor: | Chemotaxis protein cheA, Chemotaxis protein cheY | | Authors: | Park, S.Y, Beel, B.D, Simon, M.I, Bilwes, A.M, Crane, B.R. | | Deposit date: | 2004-07-14 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | In different organisms, the mode of interaction between two signaling proteins is not necessarily conserved
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1U13
 
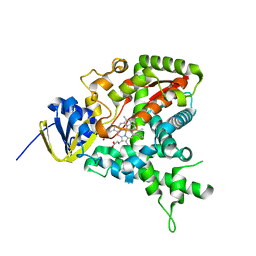 | |
8BD9
 
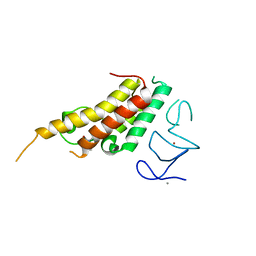 | | Crystal structure of TRIM33 alpha PHD-Bromo domain in complex with 10 | | Descriptor: | 1,3-dimethylbenzimidazol-2-one, CALCIUM ION, E3 ubiquitin-protein ligase TRIM33, ... | | Authors: | Tassone, G, Pozzi, C, Palomba, T. | | Deposit date: | 2022-10-18 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Exploiting ELIOT for Scaffold-Repurposing Opportunities: TRIM33 a Possible Novel E3 Ligase to Expand the Toolbox for PROTAC Design.
Int J Mol Sci, 23, 2022
|
|
8BGM
 
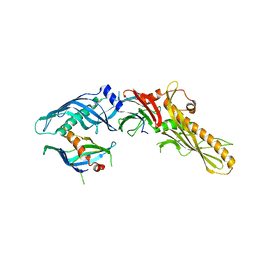 | |
1U1C
 
 | | Structure of E. coli uridine phosphorylase complexed to 5-benzylacyclouridine (BAU) | | Descriptor: | 1-((2-HYDROXYETHOXY)METHYL)-5-BENZYLPYRIMIDINE-2,4(1H,3H)-DIONE, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Bu, W, Settembre, E.C, Ealick, S.E. | | Deposit date: | 2004-07-15 | | Release date: | 2005-07-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for inhibition of Escherichia coli uridine phosphorylase by 5-substituted acyclouridines.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
8BFN
 
 | | E. coli Wadjet JetABC dimer of dimers | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, JetA, JetB, ... | | Authors: | Roisne-Hamelin, F, Beckert, B, Myasnikov, A, Li, Y, Gruber, S. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | DNA-measuring Wadjet SMC ATPases restrict smaller circular plasmids by DNA cleavage.
Mol.Cell, 82, 2022
|
|
1U3E
 
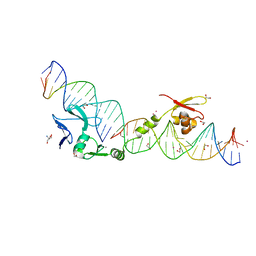 | | DNA binding and cleavage by the HNH homing endonuclease I-HmuI | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 36-MER, ... | | Authors: | Shen, B.W, Landthaler, M, Shub, D.A, Stoddard, B.L. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | DNA Binding and Cleavage by the HNH Homing Endonuclease I-HmuI.
J.Mol.Biol., 342, 2004
|
|
