7ESW
 
 | |
7ESA
 
 | | the complex structure of flavin transferase FmnB complexed with FAD | | Descriptor: | FAD:protein FMN transferase, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION | | Authors: | Zheng, Y.H, Cheng, W. | | Deposit date: | 2021-05-09 | | Release date: | 2021-11-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the catalytic and inhibitory mechanisms of the flavin transferase FmnB in Listeria monocytogenes.
MedComm (2020), 3, 2022
|
|
7ESB
 
 | | FmnB complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FAD:protein FMN transferase, MAGNESIUM ION | | Authors: | Zheng, Y.H, Cheng, W. | | Deposit date: | 2021-05-09 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the catalytic and inhibitory mechanisms of the flavin transferase FmnB in Listeria monocytogenes.
MedComm (2020), 3, 2022
|
|
7EQZ
 
 | | Crystal structure of Carboxypeptidase B complexed with Potato Carboxypeptidase Inhibitor | | Descriptor: | Carboxypeptidase B, GLYCINE, Metallocarboxypeptidase inhibitor, ... | | Authors: | Choong, Y.K, Gavor, E, Jobichen, C, Sivaraman, J. | | Deposit date: | 2021-05-05 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Aedes aegypti carboxypeptidase B1-inhibitor complex uncover the disparity between mosquito and non-mosquito insect carboxypeptidase inhibition mechanism.
Protein Sci., 30, 2021
|
|
5FQV
 
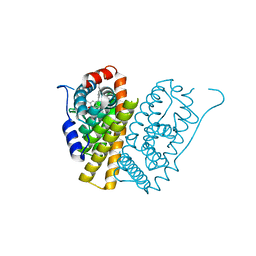 | | Selective estrogen receptor downregulator antagonists: Tetrahydroisoquinoline phenols 5. | | Descriptor: | (E)-3-[4-(6-hydroxy-2-isobutyl-7-methyl-3,4-dihydro-1H-isoquinolin-1-yl)phenyl]prop-2-enoic acid, ESTROGEN RECEPTOR ALPHA | | Authors: | Scott, J.S, bailey, A, Davies, R.D.M, Degorce, S.L, MacFaul, P.A, Gingell, H, Moss, T, Norman, R.A, Pink, J.H, Rabow, A.A, Roberts, B, Smith, P.D. | | Deposit date: | 2015-12-14 | | Release date: | 2016-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Tetrahydroisoquinoline Phenols: Selective Estrogen Receptor Downregulator Antagonists with Oral Bioavailability in Rat.
Acs Med.Chem.Lett., 7, 2016
|
|
7F2U
 
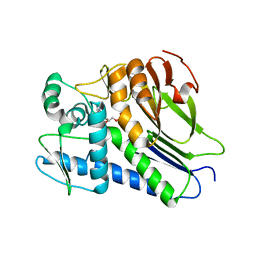 | | FmnB complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FAD:protein FMN transferase, MAGNESIUM ION | | Authors: | Cheng, W, Zheng, Y.H. | | Deposit date: | 2021-06-14 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Structural insights into the catalytic and inhibitory mechanisms of the flavin transferase FmnB in Listeria monocytogenes.
MedComm (2020), 3, 2022
|
|
7EVT
 
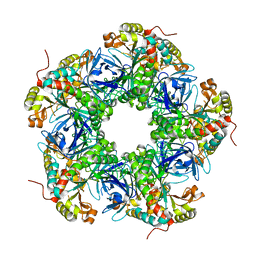 | | Crystal structure of the N-terminal degron-truncated human glutamine synthetase | | Descriptor: | Glutamine synthetase | | Authors: | Chek, M.F, Kim, S.Y, Mori, T, Hakoshima, T. | | Deposit date: | 2021-05-22 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of N-terminal degron-truncated human glutamine synthetase.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7ETK
 
 | |
4GJD
 
 | | Crystal structure of renin in complex with NVP-BGQ311 (compound 12) | | Descriptor: | (3S,5R)-N-{[9-(4-methoxybutyl)-9H-xanthen-9-yl]methyl}-5-{[(4-methylphenyl)sulfonyl]amino}piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin, ... | | Authors: | Ostermann, N, Zink, F, Kroemer, M. | | Deposit date: | 2012-08-09 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A novel class of oral direct Renin inhibitors: highly potent 3,5-disubstituted piperidines bearing a tricyclic p3-p1 pharmacophore.
J.Med.Chem., 56, 2013
|
|
4GJ8
 
 | | Crystal structure of renin in complex with PKF909-724 (compound 3) | | Descriptor: | (2R)-1-(pyrrolidin-1-yl)-3-(9H-thioxanthen-9-yl)propan-2-ol, (2S)-1-(pyrrolidin-1-yl)-3-(9H-thioxanthen-9-yl)propan-2-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ostermann, N, Zink, F, Kroemer, M. | | Deposit date: | 2012-08-09 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A novel class of oral direct Renin inhibitors: highly potent 3,5-disubstituted piperidines bearing a tricyclic p3-p1 pharmacophore.
J.Med.Chem., 56, 2013
|
|
7OTJ
 
 | | Crystal structure of Pif1 helicase from Candida albicans | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase PIF1, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Rety, S, Xi, X.G. | | Deposit date: | 2021-06-10 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural study of the function of Candida Albicans Pif1.
Biochem.Biophys.Res.Commun., 567, 2021
|
|
7P1L
 
 | | The MARK3 Kinase Domain Bound To AA-CS-1-008 | | Descriptor: | 1,2-ETHANEDIOL, 5-Bromo-4-N-[2-(1H-imidazol-5-yl)ethyl]-2-N-[3-(morpholin-4-ylmethyl)phenyl]pyrimidine-2,4-diamine, MAP/microtubule affinity-regulating kinase 3 | | Authors: | Dederer, V, Preuss, F, Chatterjee, D, Vlassova, A, Mathea, S, Axtman, A, Knapp, S. | | Deposit date: | 2021-07-01 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of Pyrimidine-Based Lead Compounds for Understudied Kinases Implicated in Driving Neurodegeneration.
J.Med.Chem., 65, 2022
|
|
6MJL
 
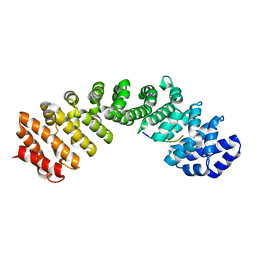 | | Crystal structure of ChREBP NLS peptide bound to importin alpha. | | Descriptor: | ChREBP Peptide ASN-TYR-TRP-LYS-ARG-ARG-ILE-GLU-VAL, Importin subunit alpha-1 | | Authors: | Jung, H, Uyeda, K. | | Deposit date: | 2018-09-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of importin alpha and the nuclear localization peptide of ChREBP, and small compound inhibitors of ChREBP-importin alpha interactions.
Biochem.J., 477, 2020
|
|
8G2J
 
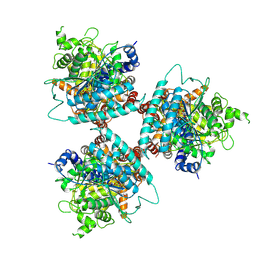 | | Hybrid aspen cellulose synthase-8 bound to UDP-glucose | | Descriptor: | Cellulose synthase, MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE-GLUCOSE, ... | | Authors: | Verma, P, Zimmer, J. | | Deposit date: | 2023-02-04 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Insights into substrate coordination and glycosyl transfer of poplar cellulose synthase-8.
Biorxiv, 2023
|
|
5EV1
 
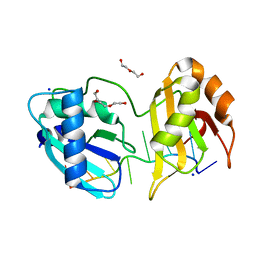 | | Structure I of Intact U2AF65 Recognizing a 3' Splice Site Signal | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA/RNA (5'-R(*UP*UP*U)-D(P*UP*UP*(BRU)P*U)-R(P*UP*U)-3'), SODIUM ION, ... | | Authors: | Agrawal, A.A, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2015-11-19 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.037 Å) | | Cite: | An extended U2AF(65)-RNA-binding domain recognizes the 3' splice site signal.
Nat Commun, 7, 2016
|
|
5EV2
 
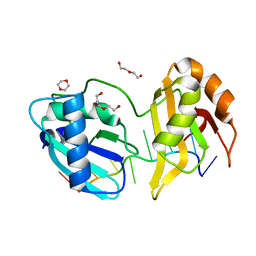 | | Structure II of Intact U2AF65 Recognizing the 3' Splice Site Signal | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DI(HYDROXYETHYL)ETHER, DNA (5'-R(P*UP*U)-D(P*UP*U)-R(P*U)-D(P*UP*(BRU)P*U)-3'), ... | | Authors: | Agrawal, A.A, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2015-11-19 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | An extended U2AF(65)-RNA-binding domain recognizes the 3' splice site signal.
Nat Commun, 7, 2016
|
|
5EV3
 
 | |
5QY3
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment F2X-Entry B03a | | Descriptor: | 2-[(1~{S})-1-azanylpropyl]phenol, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5QY2
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment F2X-Entry A12b | | Descriptor: | 3-(propan-2-yl)-1,2,4-oxadiazol-5(4H)-one, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5QY4
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment F2X-Entry B08a | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, R-1,2-PROPANEDIOL, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5QY6
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment F2X-Entry C05a | | Descriptor: | 2-methoxy-4-[[(4~{S},5~{S})-2,4,5-tris(2-methoxypyridin-4-yl)imidazolidin-1-yl]methyl]pyridine, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5QY5
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment F2X-Entry C02a | | Descriptor: | 2,4,5-tris(fluoranyl)-3-methoxy-benzoic acid, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5QY1
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment F2X-Entry A07a | | Descriptor: | (2-aminopyridin-3-yl)methanol, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5EV4
 
 | |
5QY7
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment F2X-Entry C08a | | Descriptor: | (1S,3S)-2-methyl-2,3-dihydro-1H-isoindole-1,3-diol, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
