2G9J
 
 | | Complex of TM1a(1-14)Zip with TM9a(251-284): a model for the polymerization domain ("overlap region") of tropomyosin, Northeast Structural Genomics Target OR9 | | Descriptor: | Tropomyosin 1 alpha chain, Tropomyosin 1 alpha chain/General control protein GCN4 | | Authors: | Greenfield, N.J, Huang, Y.J, Swapna, G.V.T, Bhattacharya, A, Singh, A, Montelione, G.T, Hitchcock-DeGregori, S.E, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-03-06 | | Release date: | 2006-11-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Junction between Tropomyosin Molecules: Implications for Actin Binding and Regulation.
J.Mol.Biol., 364, 2006
|
|
8U0G
 
 | |
8U1J
 
 | |
8UVX
 
 | | CosR DNA bound form I | | Descriptor: | DNA (5'-D(*AP*TP*AP*TP*CP*TP*TP*AP*AP*TP*TP*TP*TP*GP*GP*TP*TP*AP*AP*TP*A)-3'), DNA (5'-D(*TP*AP*TP*TP*AP*AP*CP*CP*AP*AP*AP*AP*TP*TP*AP*AP*GP*AP*TP*AP*T)-3'), DNA-binding response regulator | | Authors: | Zhang, Z. | | Deposit date: | 2023-11-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of DNA recognition of the Campylobacter jejuni CosR regulator.
Mbio, 15, 2024
|
|
4R9S
 
 | | Mycobacterium tuberculosis InhA bound to NITD-916 | | Descriptor: | 6-[(4,4-dimethylcyclohexyl)methyl]-4-hydroxy-3-phenylpyridin-2(1H)-one, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Noble, C.G. | | Deposit date: | 2014-09-07 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Direct inhibitors of InhA are active against Mycobacterium tuberculosis
Sci Transl Med, 7, 2015
|
|
3NYU
 
 | |
2KLJ
 
 | | Solution Structure of gammaD-Crystallin with RDC and SAXS | | Descriptor: | Gamma-crystallin D | | Authors: | Wang, J, Zuo, X, Yu, P, Byeon, I, Jung, J, Gronenborn, A.M, Wang, Y. | | Deposit date: | 2009-07-06 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Determination of multicomponent protein structures in solution using global orientation and shape restraints.
J.Am.Chem.Soc., 131, 2009
|
|
2F74
 
 | | Murine MHC class I H-2Db in complex with human b2-microglobulin and LCMV-derived immunodminant peptide gp33 | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Achour, A, Michaelsson, J, Harris, R.A, Ljunggren, H.G, Karre, K, Schneider, G, Sandalova, T. | | Deposit date: | 2005-11-30 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of the Differential Stability and Receptor Specificity of H-2D(b) in Complex with Murine versus Human beta(2)-Microglobulin.
J.Mol.Biol., 356, 2006
|
|
3DKU
 
 | | Crystal structure of Nudix hydrolase Orf153, ymfB, from Escherichia coli K-1 | | Descriptor: | Putative phosphohydrolase | | Authors: | Hong, M.K, Kim, J.K, Jung, J.H, Jung, J.W, Choi, J.Y, Kang, L.W. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of Nudix hydrolase Orf153, ymfB, from Escherichia coli K-1.
To be Published
|
|
2M3G
 
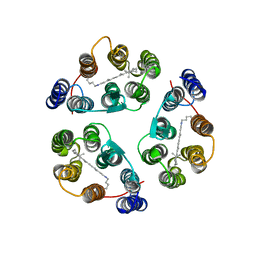 | | Structure of Anabaena Sensory Rhodopsin Determined by Solid State NMR Spectroscopy | | Descriptor: | Anabaena Sensory Rhodopsin, RETINAL | | Authors: | Wang, S, Munro, R.A, Shi, L, Kawamura, I, Okitsu, T, Wada, A, Kim, S, Jung, K, Brown, L.S, Ladizhansky, V. | | Deposit date: | 2013-01-17 | | Release date: | 2013-08-21 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | Solid-state NMR spectroscopy structure determination of a lipid-embedded heptahelical membrane protein.
Nat.Methods, 10, 2013
|
|
1J90
 
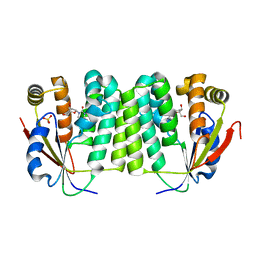 | | Crystal Structure of Drosophila Deoxyribonucleoside Kinase | | Descriptor: | 2'-DEOXYCYTIDINE, Deoxyribonucleoside kinase, SULFATE ION | | Authors: | Johansson, K, Ramaswamy, S, Ljungkrantz, C, Knecht, W, Piskur, J, Munch-Petersen, B, Eriksson, S, Eklund, H. | | Deposit date: | 2001-05-23 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural basis for substrate specificities of cellular deoxyribonucleoside kinases.
Nat.Struct.Biol., 8, 2001
|
|
1GJ2
 
 | | CO(III)-BLEOMYCIN-OOH BOUND TO AN OLIGONUCLEOTIDE CONTAINING A PHOSPHOGLYCOLATE LESION | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, 5'-D(*CP*CP*AP*AP*AP*G)-3', 5'-D(*CP*CP*CP*AP*GP*TP*AP*CP*TP*TP*TP*GP*G)-3', ... | | Authors: | Hoehn, S.T, Junker, H.-D, Bunt, R.C, Turner, C.J, Stubbe, J. | | Deposit date: | 2000-11-01 | | Release date: | 2001-06-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Co(III)-bleomycin-OOH bound to a phosphoglycolate lesion containing oligonucleotide: implications for bleomycin-induced double-strand DNA cleavage.
Biochemistry, 40, 2001
|
|
1G5L
 
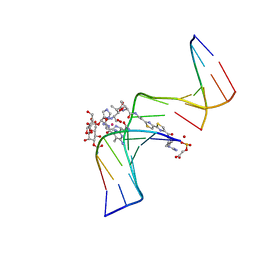 | | CO(III)-BLEOMYCIN-OOH BOUND TO AN OLIGONUCLEOTIDE CONTAINING A PHOSPHOGLYCOLATE LESION | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, 5'-D(*CP*CP*AP*AP*AP*G)-3', 5'-D(*CP*CP*CP*AP*GP*TP*AP*CP*TP*TP*TP*GP*G)-3', ... | | Authors: | Hoehn, S.T, Junker, H.-D, Bunt, R.C, Turner, C.J, Stubbe, J. | | Deposit date: | 2000-11-01 | | Release date: | 2001-06-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Co(III)-bleomycin-OOH bound to a phosphoglycolate lesion containing oligonucleotide: implications for bleomycin-induced double-strand DNA cleavage.
Biochemistry, 40, 2001
|
|
2N50
 
 | | Novel Structural Components Contribute to the High Thermal Stability of Acyl Carrier Protein from Enterococcus faecalis | | Descriptor: | Acyl carrier protein | | Authors: | Park, Y, Jung, M, Song, H, Jeong, K, Kim, Y. | | Deposit date: | 2015-07-02 | | Release date: | 2015-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Novel Structural Components Contribute to the High Thermal Stability of Acyl Carrier Protein from Enterococcus faecalis.
J. Biol. Chem., 291, 2016
|
|
3AAG
 
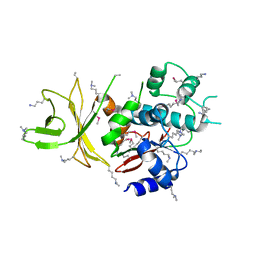 | | Crystal structure of C. jejuni pglb C-terminal domain | | Descriptor: | CALCIUM ION, General glycosylation pathway protein | | Authors: | Maita, N, Kohda, D. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Comparative structural biology of Eubacterial and Archaeal oligosaccharyltransferases.
J.Biol.Chem., 285, 2010
|
|
3N0I
 
 | | Crystal Structure of Ad37 fiber knob in complex with GD1a oligosaccharide | | Descriptor: | Fiber, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ZINC ION | | Authors: | Nilsson, E.C, Storm, R.J, Bauer, J, Johansson, S.M.C, Lookene, A, Angstroem, J, Hedenstroem, M, Fraengsmyr, L, Rinaldi, S, Willison, H, Domelloef, F.P, Stehle, T, Arnberg, N. | | Deposit date: | 2010-05-14 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The GD1a glycan is a cellular receptor for adenoviruses causing epidemic keratoconjunctivitis.
NAT.MED. (N.Y.), 17, 2011
|
|
2HBS
 
 | | THE HIGH RESOLUTION CRYSTAL STRUCTURE OF DEOXYHEMOGLOBIN S | | Descriptor: | HEMOGLOBIN S (DEOXY), ALPHA CHAIN, BETA CHAIN, ... | | Authors: | Harrington, D.J, Adachi, K, Royer Junior, W.E. | | Deposit date: | 1997-05-06 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The high resolution crystal structure of deoxyhemoglobin S.
J.Mol.Biol., 272, 1997
|
|
2OEP
 
 | | MSrecA-ADP-complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein recA | | Authors: | Krishna, R, Rajan Prabu, J, Manjunath, G.P, Datta, S, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2006-12-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Snapshots of RecA protein involving movement of the C-domain and different conformations of the DNA-binding loops: crystallographic and comparative analysis of 11 structures of Mycobacterium smegmatis RecA
J.Mol.Biol., 367, 2007
|
|
2OFO
 
 | | MSrecA-native | | Descriptor: | PHOSPHATE ION, Protein recA | | Authors: | Krishna, R, Rajan Prabu, J, Manjunath, G.P, Datta, S, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2007-01-04 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Snapshots of RecA protein involving movement of the C-domain and different conformations of the DNA-binding loops: crystallographic and comparative analysis of 11 structures of Mycobacterium smegmatis RecA
J.Mol.Biol., 367, 2007
|
|
2ODW
 
 | | MSrecA-ATP-GAMA-S complex | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Protein recA | | Authors: | Krishna, R, Rajan Prabu, J, Manjunath, G.P, Datta, S, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2006-12-27 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Snapshots of RecA protein involving movement of the C-domain and different conformations of the DNA-binding loops: crystallographic and comparative analysis of 11 structures of Mycobacterium smegmatis RecA
J.Mol.Biol., 367, 2007
|
|
2ODN
 
 | | MSRECA-dATP complex | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Protein recA | | Authors: | Krishna, R, Rajan Prabu, J, Manjunath, G.P, Datta, S, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2006-12-24 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Snapshots of RecA protein involving movement of the C-domain and different conformations of the DNA-binding loops: crystallographic and comparative analysis of 11 structures of Mycobacterium smegmatis RecA
J.Mol.Biol., 367, 2007
|
|
2OE2
 
 | | MSrecA-native-low humidity 95% | | Descriptor: | PHOSPHATE ION, Protein recA | | Authors: | Krishna, R, Rajan Prabu, J, Manjunath, G.P, Datta, S, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2006-12-28 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Snapshots of RecA protein involving movement of the C-domain and different conformations of the DNA-binding loops: crystallographic and comparative analysis of 11 structures of Mycobacterium smegmatis RecA
J.Mol.Biol., 367, 2007
|
|
2I22
 
 | | Crystal structure of Escherichia coli phosphoheptose isomerase in complex with reaction substrate sedoheptulose 7-phosphate | | Descriptor: | D-ALTRO-HEPT-2-ULOSE 7-PHOSPHATE, Phosphoheptose isomerase | | Authors: | Blakely, K, Zhang, K, DeLeon, G, Wright, G, Junop, M. | | Deposit date: | 2006-08-15 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and Function of Sedoheptulose-7-phosphate Isomerase, a Critical Enzyme for Lipopolysaccharide Biosynthesis and a Target for Antibiotic Adjuvants
J.Biol.Chem., 283, 2008
|
|
2I2W
 
 | | Crystal Structure of Escherichia Coli Phosphoheptose Isomerase | | Descriptor: | GLYCEROL, Phosphoheptose isomerase | | Authors: | DeLeon, G, Blakely, K, Zhang, K, Wright, G, Junop, M. | | Deposit date: | 2006-08-17 | | Release date: | 2007-08-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Function of Sedoheptulose-7-phosphate Isomerase, a Critical Enzyme for Lipopolysaccharide Biosynthesis and a Target for Antibiotic Adjuvants
J.Biol.Chem., 283, 2008
|
|
2OES
 
 | | MSrecA-native-SSB | | Descriptor: | PHOSPHATE ION, Protein recA | | Authors: | Krishna, R, Rajan Prabu, J, Manjunath, G.P, Datta, S, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2007-01-01 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Snapshots of RecA protein involving movement of the C-domain and different conformations of the DNA-binding loops: crystallographic and comparative analysis of 11 structures of Mycobacterium smegmatis RecA
J.Mol.Biol., 367, 2007
|
|
