6I1F
 
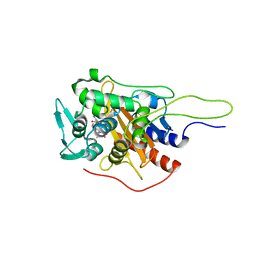 | | Crystal structure of TP domain from Chlamydia trachomatis Penicillin-Binding Protein 3 in complex with amoxicillin | | Descriptor: | 2-{1-[2-AMINO-2-(4-HYDROXY-PHENYL)-ACETYLAMINO]-2-OXO-ETHYL}-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, Penicillin-binding protein,Penicillin-binding protein | | Authors: | Bellini, D, Koekemoer, L, Newman, H, Dowson, C.G. | | Deposit date: | 2018-10-28 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of TP domain from Chlamydia trachomatis Penicillin-Binding Protein 3 in complex with amoxicillin
To Be Published
|
|
6I1H
 
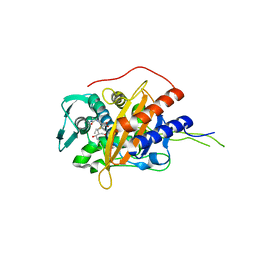 | | Crystal structure of TP domain from Chlamydia trachomatis Penicillin-Binding Protein 3 in complex with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein,Penicillin-binding protein | | Authors: | Bellini, D, Koekemoer, L, Newman, H, Dowson, C.G. | | Deposit date: | 2018-10-28 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of TP domain from Chlamydia trachomatis Penicillin-Binding Protein 3 in complex with meropenem
To Be Published
|
|
4P85
 
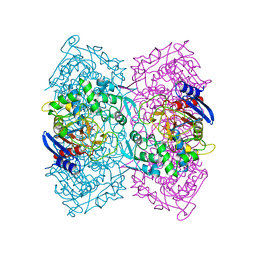 | | Crystal structure of Est-Y29, a novel penicillin-binding protein/beta-lactamase homolog from a metagenomic library | | Descriptor: | DIETHYL PHOSPHONATE, Est-Y29 | | Authors: | Ngo, T.D, Ryu, B.H, Ju, H.S, Jang, E.J, Kim, K.K, Kim, D.H. | | Deposit date: | 2014-03-30 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic analysis and biochemical applications of a novel penicillin-binding protein/ beta-lactamase homologue from a metagenomic library.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4P87
 
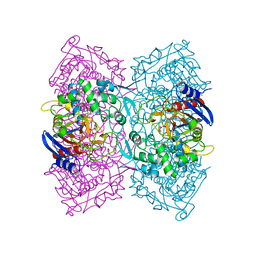 | | Crystal structure of Est-Y29, a novel penicillin-binding protein/beta-lactamase homolog from a metagenomic library | | Descriptor: | 4-NITROPHENYL PHOSPHATE, Est-Y29 | | Authors: | Ngo, T.D, Ryu, B.H, Ju, H.S, Jang, E.J, Kim, K.K, Kim, D.H. | | Deposit date: | 2014-03-30 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Crystallographic analysis and biochemical applications of a novel penicillin-binding protein/ beta-lactamase homologue from a metagenomic library.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1Y1O
 
 | | X-ray crystal Structure of Penicillin-binding protein-related factor A from Bacillus stearothermophilus | | Descriptor: | NICKEL (II) ION, Penicillin-binding protein-related factor A, SULFATE ION | | Authors: | Osipiuk, J, Li, H, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-11-19 | | Release date: | 2004-12-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystal Structure of Penicillin-binding protein-related factor A from Bacillus stearothermophilus
To be Published
|
|
7U4H
 
 | |
4P6B
 
 | | Crystal structure of Est-Y29,a novel penicillin-binding protein/beta-lactamase homolog from a metagenomic library | | Descriptor: | Est-Y29 | | Authors: | Ngo, T.D, Ryu, B.H, Ju, H.S, Jang, E.J, Kim, K.K, Kim, D.H. | | Deposit date: | 2014-03-24 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic analysis and biochemical applications of a novel penicillin-binding protein/ beta-lactamase homologue from a metagenomic library.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6MKH
 
 | | Crystal structure of pencillin binding protein 4 (PBP4) from Enterococcus faecalis in the imipenem-bound form | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, PHOSPHATE ION, pencillin binding protein 4 (PBP4) | | Authors: | D'Andrea, E.D, Moon, T.M, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6MKF
 
 | | Crystal structure of penicillin binding protein 5 (PBP5) from Enterococcus faecium in the imipenem-bound form | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, SULFATE ION, penicillin binding protein 5 (PBP5) | | Authors: | Moon, T.M, Lee, C, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6MKG
 
 | | Crystal structure of penicillin binding protein 5 (PBP5) from Enterococcus faecium in the benzylpenicilin-bound form | | Descriptor: | OPEN FORM - PENICILLIN G, SULFATE ION, penicillin binding protein 5 (PBP5) | | Authors: | Moon, T.M, Lee, C, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6MKJ
 
 | | Crystal structure of penicillin binding protein 5 (PBP5) from Enterococcus faecium in the closed conformation | | Descriptor: | penicillin binding protein 5 (PBP5) | | Authors: | Moon, T.M, Soares, A, D'Andrea, E.D, Jaconcic, J, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.864 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6MKA
 
 | | Crystal structure of penicillin binding protein 5 (PBP5) from Enterococcus faecium in the open conformation | | Descriptor: | SULFATE ION, penicillin binding protein 5 (PBP5) | | Authors: | Moon, T.M, Lee, C, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
4BJP
 
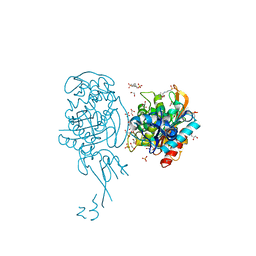 | | Crystal structure of E. coli penicillin binding protein 3 | | Descriptor: | 1,2-ETHANEDIOL, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, CHLORIDE ION, ... | | Authors: | Sauvage, E, Joris, M, Herman, R, Kerff, F, Rocaboy, M, Charlier, P. | | Deposit date: | 2013-04-19 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Penicillin-Binding Protein 3 (Pbp3) from Escherichia Coli.
Plos One, 9, 2014
|
|
4BJQ
 
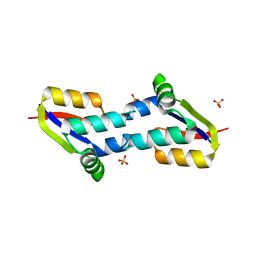 | | Crystal structure of E. coli penicillin binding protein 3, domain V88- S165 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PENICILLIN BINDING PROTEIN TRANSPEPTIDASE DOMAIN PROTEIN, SULFATE ION | | Authors: | Sauvage, E, Joris, M, Herman, R, Kerff, F, Rocaboy, M, Charlier, P. | | Deposit date: | 2013-04-19 | | Release date: | 2014-05-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Penicillin-Binding Protein 3 (Pbp3) from Escherichia Coli.
Plos One, 9, 2014
|
|
4Q6V
 
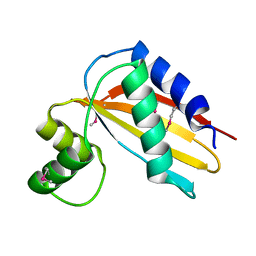 | |
4Q6Z
 
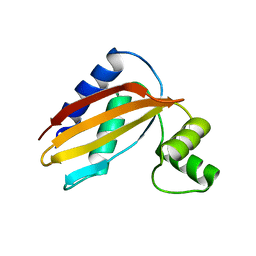 | | LpoB C-terminal domain from Escherichia coli | | Descriptor: | Penicillin-binding protein activator LpoB | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2014-04-23 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Lipoprotein Outer Membrane Regulator of Penicillin-binding Protein 1B.
J.Biol.Chem., 289, 2014
|
|
4Q6L
 
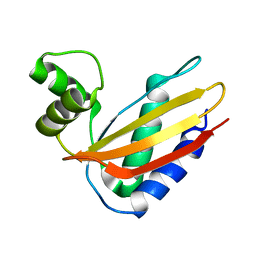 | |
5LP4
 
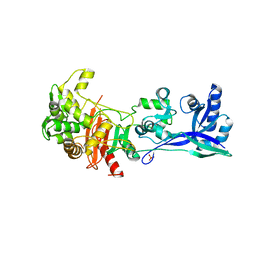 | | Penicillin-Binding Protein (PBP2) from Helicobacter pylori | | Descriptor: | Penicillin-binding protein 2 (Pbp2), SULFATE ION | | Authors: | Contreras-Martel, C, Martins, A, Ecobichon, C, Maragno, D.M, Mattei, P.J, El Ghachi, M, Boneca, I.G, Dessen, A. | | Deposit date: | 2016-08-11 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Molecular architecture of the PBP2-MreC core bacterial cell wall synthesis complex.
Nat Commun, 8, 2017
|
|
1O75
 
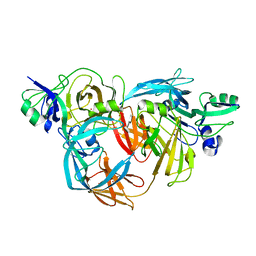 | | Tp47, the 47-Kilodalton Lipoprotein of Treponema pallidum | | Descriptor: | 2,3-di-O-sulfo-alpha-D-glucopyranose-(1-6)-2,3-di-O-sulfo-alpha-D-glucopyranose, 47 KDA MEMBRANE ANTIGEN, XENON | | Authors: | Deka, R.K, Machius, M, Norgard, M.V, Tomchick, D.R. | | Deposit date: | 2002-10-23 | | Release date: | 2002-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the 47-kDa lipoprotein of Treponema pallidum reveals a novel penicillin-binding protein.
J. Biol. Chem., 277, 2002
|
|
4KQQ
 
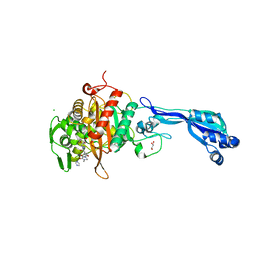 | | CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 3 FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH (5S)-Penicilloic Acid | | Descriptor: | (2S,4S)-2-[(R)-carboxy{[(2R)-2-{[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]amino}-2-phenylacetyl]amino}methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
4KQR
 
 | | CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 3 FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH (5S)-Penicilloic Acid | | Descriptor: | (2S,4S)-2-[(R)-carboxy{[(2R)-2-{[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]amino}-2-phenylacetyl]amino}methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
4KQO
 
 | | Crystal structure of penicillin-binding protein 3 from pseudomonas aeruginosa in complex with piperacillin | | Descriptor: | CHLORIDE ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
6C39
 
 | |
1TVF
 
 | | Crystal Structure of penicillin-binding protein 4 (PBP4) from Staphylococcus aureus | | Descriptor: | SULFATE ION, UNKNOWN LIGAND, penicillin binding protein 4 | | Authors: | Rajashankar, K.R, Ray, S.S, Bonanno, J.B, Pinho, M, Tomasz, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-29 | | Release date: | 2004-07-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of penicillin-binding protein 4 (PBP4) from Staphylococcus aureus
To be Published
|
|
2J8Y
 
 | | Structure of PBP-A acyl-enzyme complex with penicillin-G | | Descriptor: | OPEN FORM - PENICILLIN G, TLL2115 PROTEIN | | Authors: | Evrard, C, Declercq, J.P. | | Deposit date: | 2006-10-31 | | Release date: | 2007-11-27 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Pbp-A from Thermosynechococcus Elongatus, a Penicillin-Binding Protein Closely Related to Class a Beta-Lactamases.
J.Mol.Biol., 386, 2009
|
|
