8WOU
 
 | |
4H51
 
 | |
6C9B
 
 | | The structure of MppP soaked with the products 4HKA and 2KA | | Descriptor: | (4S)-5-carbamimidamido-4-hydroxy-2-oxopentanoic acid, CHLORIDE ION, PLP-Dependent L-Arginine Hydroxylase MppP | | Authors: | Han, L, Silvaggi, N.R. | | Deposit date: | 2018-01-26 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.689 Å) | | Cite: | Streptomyces wadayamensis MppP is a PLP-Dependent Oxidase, Not an Oxygenase.
Biochemistry, 57, 2018
|
|
3QPG
 
 | |
3QN6
 
 | |
9AAT
 
 | |
7BXS
 
 | | 2-amino-3-ketobutyrate CoA ligase from Cupriavidus necator glycine binding form | | Descriptor: | 2-amino-3-ketobutyrate coenzyme A ligase, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE] | | Authors: | Motoyama, T, Nakano, S, Hasebe, F, Miyoshi, N, Ito, S. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Chemoenzymatic synthesis of 3-ethyl-2,5-dimethylpyrazine by L-threonine 3-dehydrogenase and 2-amino-3-ketobutyrate CoA ligase/L-threonine aldolase
Commun Chem, 4, 2021
|
|
7BXQ
 
 | | 2-amino-3-ketobutyrate CoA ligase from Cupriavidus necator L-Threonine binding form | | Descriptor: | 2-amino-3-ketobutyrate coenzyme A ligase, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-allothreonine | | Authors: | Motoyama, T, Nakano, S, Hasebe, F, Miyoshi, N, Ito, S. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Chemoenzymatic synthesis of 3-ethyl-2,5-dimethylpyrazine by L-threonine 3-dehydrogenase and 2-amino-3-ketobutyrate CoA ligase/L-threonine aldolase
Commun Chem, 4, 2021
|
|
7BXP
 
 | | 2-amino-3-ketobutyrate CoA ligase from Cupriavidus necator | | Descriptor: | 2-amino-3-ketobutyrate coenzyme A ligase | | Authors: | Motoyama, T, Nakano, S, Hasebe, F, Miyoshi, N, Ito, S. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chemoenzymatic synthesis of 3-ethyl-2,5-dimethylpyrazine by L-threonine 3-dehydrogenase and 2-amino-3-ketobutyrate CoA ligase/L-threonine aldolase
Commun Chem, 4, 2021
|
|
7BXR
 
 | | 2-amino-3-ketobutyrate CoA ligase from Cupriavidus necator 3-Hydroxynorvaline binding form | | Descriptor: | (2S,3R)-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-3-oxidanyl-pentanoic acid, 2-amino-3-ketobutyrate coenzyme A ligase | | Authors: | Motoyama, T, Nakano, S, Hasebe, F, Miyoshi, N, Ito, S. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Chemoenzymatic synthesis of 3-ethyl-2,5-dimethylpyrazine by L-threonine 3-dehydrogenase and 2-amino-3-ketobutyrate CoA ligase/L-threonine aldolase
Commun Chem, 4, 2021
|
|
4IW7
 
 | | Crystal structure of 8-amino-7-oxononanoate synthase (bioF) from Francisella tularensis. | | Descriptor: | 8-amino-7-oxononanoate synthase | | Authors: | Newcomb, W, Niedzialkowska, E, Porebski, P.J, Grimshaw, S, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-23 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of 8-amino-7-oxononanoate synthase (bioF) from Francisella tularensis.
To be Published
|
|
8XHK
 
 | | Crystal structure of alpha-Oxoamine Synthase Alb29 with PLP cofactor | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Xu, M.J, Zhang, D.K. | | Deposit date: | 2023-12-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural and mechanistic investigations on CC bond forming alpha-oxoamine synthase allowing L-glutamate as substrate.
Int.J.Biol.Macromol., 268, 2024
|
|
8XHA
 
 | | Crystal structure of alpha-Oxoamine Synthase Alb29 with PLP cofactor and L-glutamate | | Descriptor: | 8-amino-7-oxononanoate synthase, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-glutamic acid, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Xu, M.J, Zhang, D.K. | | Deposit date: | 2023-12-17 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural and mechanistic investigations on CC bond forming alpha-oxoamine synthase allowing L-glutamate as substrate.
Int.J.Biol.Macromol., 268, 2024
|
|
8XHD
 
 | | Crystal structure of alpha-Oxoamine Synthase Alb29 with PLP cofactor and L-glutamate | | Descriptor: | 8-amino-7-oxononanoate synthase, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-glutamic acid, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Xu, M.J, Zhang, D.K. | | Deposit date: | 2023-12-17 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and mechanistic investigations on CC bond forming alpha-oxoamine synthase allowing L-glutamate as substrate.
Int.J.Biol.Macromol., 268, 2024
|
|
4IX8
 
 | |
3QGU
 
 | | L,L-Diaminopimelate aminotransferase from Chlamydomonas reinhardtii | | Descriptor: | AZIDE ION, GLYCEROL, LL-diaminopimelate aminotransferase, ... | | Authors: | Dobson, R.C.J, Giron, I, Hudson, A.O. | | Deposit date: | 2011-01-25 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | L,L-Diaminopimelate Aminotransferase from Chlamydomonas reinhardtii: A Target for Algaecide Development
Plos One, 6, 2011
|
|
6C3B
 
 | | O2-, PLP-Dependent L-Arginine Hydroxylase RohP Holoenzyme | | Descriptor: | 1,2-ETHANEDIOL, TRIETHYLENE GLYCOL, Uncharacterized protein | | Authors: | Hedges, J.B, Ryan, K.S. | | Deposit date: | 2018-01-09 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Snapshots of the Catalytic Cycle of an O2, Pyridoxal Phosphate-Dependent Hydroxylase.
ACS Chem. Biol., 13, 2018
|
|
3TCM
 
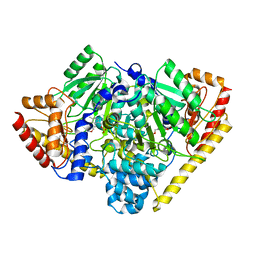 | | Crystal Structure of Alanine Aminotransferase from Hordeum vulgare | | Descriptor: | Alanine aminotransferase 2, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE | | Authors: | Rydel, T.J, Sturman, E.J, Halls, C, Chen, S, Zeng, J, Evdokimov, A, Duff, S.M.G. | | Deposit date: | 2011-08-09 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The Enzymology of alanine aminotransferase (AlaAT) isoforms from Hordeum vulgare and other organisms, and the HvAlaAT crystal structure.
Arch.Biochem.Biophys., 528, 2012
|
|
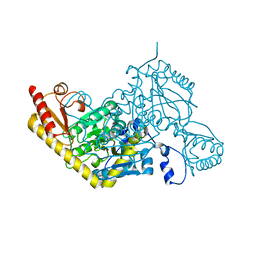
6D0A
 
 | |
3TAT
 
 | | TYROSINE AMINOTRANSFERASE FROM E. COLI | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, TYROSINE AMINOTRANSFERASE | | Authors: | Ko, T.P, Yang, W.Z, Wu, S.P, Tsai, H, Yuan, H.S. | | Deposit date: | 1998-08-12 | | Release date: | 1999-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystallization and preliminary crystallographic analysis of the Escherichia coli tyrosine aminotransferase.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
3T32
 
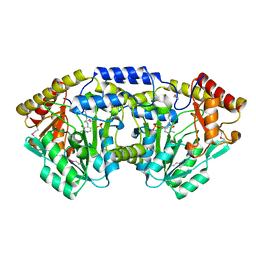 | | Crystal structure of a putative C-S lyase from Bacillus anthracis | | Descriptor: | Aminotransferase, class I/II | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Peterson, S.N, Porebski, P, Minor, W, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-24 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative C-S lyase from Bacillus anthracis
TO BE PUBLISHED
|
|
4MY5
 
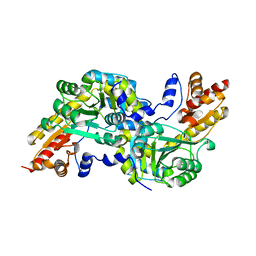 | | Crystal structure of the aromatic amino acid aminotransferase from Streptococcus mutants | | Descriptor: | Putative amino acid aminotransferase | | Authors: | Cong, X, Li, X, Ge, J, Feng, Y, Feng, X, Li, S. | | Deposit date: | 2013-09-27 | | Release date: | 2014-10-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Crystal structure of the aromatic-amino-acid aminotransferase from Streptococcus mutans.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
3TQX
 
 | | Structure of the 2-amino-3-ketobutyrate coenzyme A ligase (kbl) from Coxiella burnetii | | Descriptor: | 2-amino-3-ketobutyrate coenzyme A ligase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
2O0R
 
 | | The three-dimensional structure of N-Succinyldiaminopimelate aminotransferase from Mycobacterium tuberculosis | | Descriptor: | CHLORIDE ION, GLYCEROL, Rv0858c (N-Succinyldiaminopimelate aminotransferase), ... | | Authors: | Weyand, S, Kefala, G, Weiss, M.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-11-28 | | Release date: | 2007-02-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Three-dimensional Structure of N-Succinyldiaminopimelate Aminotransferase from Mycobacterium tuberculosis
J.Mol.Biol., 367, 2007
|
|
8AAT
 
 | |
