1U76
 
 | |
1UNB
 
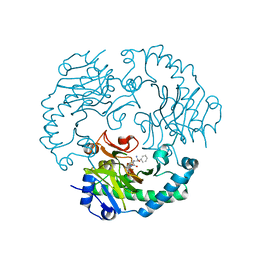 | | Deacetoxycephalosporin C synthase complexed with 2-oxoglutarate and ampicillin | | Descriptor: | (2S,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, 2-OXOGLUTARIC ACID, DEACETOXYCEPHALOSPORIN C SYNTHETASE, ... | | Authors: | Valegard, K, Terwisscha van Scheltinga, A.C, Dubus, A, Oster, L.M, Rhangino, G, Hajdu, J, Andersson, I. | | Deposit date: | 2003-09-09 | | Release date: | 2004-01-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structural Basis of Cephalosporin Formation in a Mononuclear Ferrous Enzyme
Nat.Struct.Mol.Biol., 11, 2004
|
|
1UZ3
 
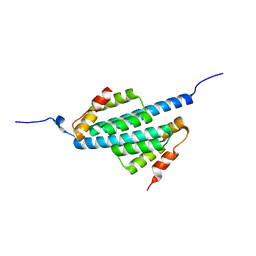 | |
1VSO
 
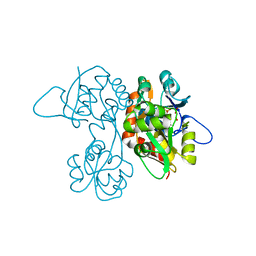 | | Crystal Structure of the Ligand-Binding Core of iGluR5 in Complex With the Antagonist (S)-ATPO at 1.85 A resolution | | Descriptor: | (S)-2-AMINO-3-(5-TERT-BUTYL-3-(PHOSPHONOMETHOXY)-4-ISOXAZOLYL)PROPIONIC ACID, GLYCEROL, Glutamate receptor, ... | | Authors: | Hald, H, Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-03-29 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Partial agonism and antagonism of the ionotropic glutamate receptor iGLuR5: structures of the ligand-binding core in complex with domoic acid and 2-amino-3-[5-tert-butyl-3-(phosphonomethoxy)-4-isoxazolyl]propionic acid.
J.Biol.Chem., 282, 2007
|
|
1W80
 
 | | Crystal structure of the alpha-adaptin appendage domain, from the AP2 adaptor complex, bound to 2 peptides from Synaptojanin170 | | Descriptor: | ADAPTER-RELATED PROTEIN COMPLEX 2 ALPHA 2 SUBUNIT, BENZAMIDINE, CARBONATE ION, ... | | Authors: | Ford, M.G.J, Praefcke, G.J.K, McMahon, H.T. | | Deposit date: | 2004-09-14 | | Release date: | 2004-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolving Nature of the Ap2 Alpha-Appendage Hub During Clathrin-Coated Vesicle Endocytosis.
Embo J., 23, 2004
|
|
1VJ3
 
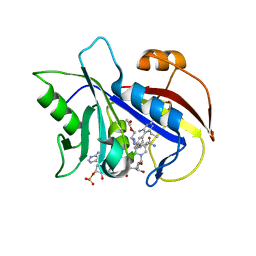 | | STRUCTURAL STUDIES ON BIO-ACTIVE COMPOUNDS. CRYSTAL STRUCTURE AND MOLECULAR MODELING STUDIES ON THE PNEUMOCYSTIS CARINII DIHYDROFOLATE REDUCTASE COFACTOR COMPLEX WITH TAB, A HIGHLY SELECTIVE ANTIFOLATE. | | Descriptor: | ACETIC ACID N-[2-CHLORO-5-[6-ETHYL-2,4-DIAMINO-PYRIMID-5-YL]-PHENYL]-[BENZYL-TRIAZEN-3-YL]ETHYL ESTER, DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Rak, D, Luft, J.R, Queener, S.F, Laughton, C.A, Malcolm, F.G. | | Deposit date: | 2004-01-13 | | Release date: | 2004-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural studies on bioactive compounds. 30. Crystal structure and molecular modeling studies on the Pneumocystis carinii dihydrofolate reductase cofactor complex with TAB, a highly selective antifolate.
Biochemistry, 39, 2000
|
|
7BM0
 
 | | Crystal structure of the tick-borne encephalitis virus NS3 helicase in complex with AMPPNP | | Descriptor: | MANGANESE (II) ION, NS3 helicase domain, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Anindita, P.D, Grinkevich, P, Franta, Z. | | Deposit date: | 2021-01-19 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insight into the RNA-stimulated ATPase activity of tick-borne encephalitis virus helicase.
J.Biol.Chem., 298, 2022
|
|
6E18
 
 | | Crystal structure of Chlamydomonas reinhardtii HAP2 ectodomain provides structural insights of functional loops in green algae. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Baquero, E, Legrand, P, Rey, F.A. | | Deposit date: | 2018-07-09 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Species-Specific Functional Regions of the Green Alga Gamete Fusion Protein HAP2 Revealed by Structural Studies.
Structure, 27, 2019
|
|
5TAS
 
 | | Structure of rabbit RyR1 (Caffeine/ATP/EGTA dataset, class 1) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5T7T
 
 | | Galectin-8 N terminal domain in complex with LNT | | Descriptor: | CHLORIDE ION, Galectin-8, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bohari, M.H, Yu, X, Blanchard, H. | | Deposit date: | 2016-09-05 | | Release date: | 2017-01-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-based rationale for differential recognition of lacto- and neolacto- series glycosphingolipids by the N-terminal domain of human galectin-8.
Sci Rep, 6, 2016
|
|
5TA3
 
 | | Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 2) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-09 | | Release date: | 2016-10-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TB2
 
 | | Structure of rabbit RyR1 (EGTA-only dataset, class 2) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-11 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TF9
 
 | |
5TAN
 
 | | Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 3) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TB3
 
 | | Structure of rabbit RyR1 (EGTA-only dataset, class 3) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-11 | | Release date: | 2016-10-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5LFF
 
 | | NMR structure of peptide 2 targeting CXCR4 | | Descriptor: | ARG-ALA-CYS-ARG-PHE-PHE-CYS | | Authors: | Di Maro, S, Trotta, A.M, Brancaccio, D, Di Leva, F.S, La Pietra, V, Ierano, C, Napolitano, M, Portella, L, D'Alterio, C, Siciliano, R.A, Sementa, D, Tomassi, S, Carotenuto, A, Novellino, E, Scala, S, Marinelli, L. | | Deposit date: | 2016-07-01 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Exploring the N-Terminal Region of C-X-C Motif Chemokine 12 (CXCL12): Identification of Plasma-Stable Cyclic Peptides As Novel, Potent C-X-C Chemokine Receptor Type 4 (CXCR4) Antagonists.
J.Med.Chem., 59, 2016
|
|
5UAG
 
 | | Escherichia coli RNA polymerase mutant - RpoB D516V | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Scharf, N.T, Stefan, M.A, Garcia, G.A, Murakami, K.S. | | Deposit date: | 2016-12-19 | | Release date: | 2017-02-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.399 Å) | | Cite: | Structural basis for rifamycin resistance of bacterial RNA polymerase by the three most clinically important RpoB mutations found in Mycobacterium tuberculosis.
Mol. Microbiol., 103, 2017
|
|
7DRN
 
 | | Structure of ATP-grasp ligase PsnB complexed with precursor peptide PsnA2 and AMPPNP | | Descriptor: | ATP-grasp domain-containing protein, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PsnA214-38, ... | | Authors: | Song, I, Yu, J, Song, W, Kim, S. | | Deposit date: | 2020-12-29 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Molecular mechanism underlying substrate recognition of the peptide macrocyclase PsnB.
Nat.Chem.Biol., 17, 2021
|
|
5MM4
 
 | |
5UA6
 
 | | Ocellatin-LB1, solution structure in SDS micelle by NMR spectroscopy | | Descriptor: | Ocellatin-LB1 | | Authors: | Gusmao, K.A.G, dos Santos, D.M, Santos, V.M, Pilo-Veloso, D, de Lima, M.E, Resende, J.M. | | Deposit date: | 2016-12-19 | | Release date: | 2017-03-29 | | Last modified: | 2018-04-18 | | Method: | SOLUTION NMR | | Cite: | NMR structures in different membrane environments of three ocellatin peptides isolated from Leptodactylus labyrinthicus.
Peptides, 103, 2018
|
|
5UAQ
 
 | | Escherichia coli RNA polymerase RpoB H526Y mutant | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Scharf, N.T, Stefan, M.A, Garcia, G.A, Murakami, K.S. | | Deposit date: | 2016-12-19 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for rifamycin resistance of bacterial RNA polymerase by the three most clinically important RpoB mutations found in Mycobacterium tuberculosis.
Mol. Microbiol., 103, 2017
|
|
5U9R
 
 | | Ocellatin-LB2, solution structure in TFE by NMR spectroscopy | | Descriptor: | Ocellatin-LB2 | | Authors: | Gusmao, K.A.G, dos Santos, D.M, Santos, V.M, Pilo-Veloso, D, Verly, R.M, de Lima, M.E, Resende, J.M. | | Deposit date: | 2016-12-18 | | Release date: | 2017-03-29 | | Last modified: | 2018-04-18 | | Method: | SOLUTION NMR | | Cite: | NMR structures in different membrane environments of three ocellatin peptides isolated from Leptodactylus labyrinthicus.
Peptides, 103, 2018
|
|
5UAJ
 
 | | Escherichia coli RNA polymerase RpoB S531L mutant | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Scharf, N.T, Stefan, M.A, Garcia, G.A, Murakami, K.S. | | Deposit date: | 2016-12-19 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.915 Å) | | Cite: | Structural basis for rifamycin resistance of bacterial RNA polymerase by the three most clinically important RpoB mutations found in Mycobacterium tuberculosis.
Mol. Microbiol., 103, 2017
|
|
5U9Q
 
 | | Ocellatin-LB1 | | Descriptor: | Ocellatin-LB1 | | Authors: | Gusmao, K.A.G, Santos, D.M, de Lima, M.E, Pilo-Veloso, D, Resende, J.M. | | Deposit date: | 2016-12-17 | | Release date: | 2017-12-13 | | Last modified: | 2018-04-18 | | Method: | SOLUTION NMR | | Cite: | NMR structures in different membrane environments of three ocellatin peptides isolated from Leptodactylus labyrinthicus.
Peptides, 103, 2018
|
|
5UA7
 
 | | Ocellatin-LB2, solution structure in SDS micelle by NMR spectroscopy | | Descriptor: | Ocellatin-LB2 | | Authors: | Gusmao, K.A.G, dos Santos, D.M, Santos, V.M, Pilo-Veloso, D, de Lima, M.E, Resende, J.M. | | Deposit date: | 2016-12-19 | | Release date: | 2017-03-29 | | Last modified: | 2018-04-18 | | Method: | SOLUTION NMR | | Cite: | NMR structures in different membrane environments of three ocellatin peptides isolated from Leptodactylus labyrinthicus.
Peptides, 103, 2018
|
|
