7Y0Z
 
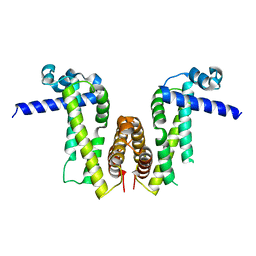 | | Crystal structure of Pseudomonas aeruginosa PvrA | | Descriptor: | TetR family transcriptional regulator | | Authors: | Liang, H, Zhang, Q, Bartlam, M. | | Deposit date: | 2022-06-06 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Regulatory and structural mechanisms of PvrA-mediated regulation of the PQS quorum-sensing system and PHA biosynthesis in Pseudomonas aeruginosa.
Nucleic Acids Res., 51, 2023
|
|
1WT1
 
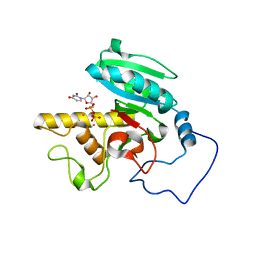 | | Mutant ABO(H) blood group glycosyltransferase with bound UDP and acceptor | | Descriptor: | Histo-blood group ABO system transferase, MERCURY (II) ION, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Lee, H.J, Barry, C.H, Borisova, S.N, Seto, N.O.L, Zheng, R.B, Blancher, A, Evans, S.V, Palcic, M.M. | | Deposit date: | 2004-11-12 | | Release date: | 2004-12-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the inactivity of human blood group o2 glycosyltransferase
J.Biol.Chem., 280, 2005
|
|
1R80
 
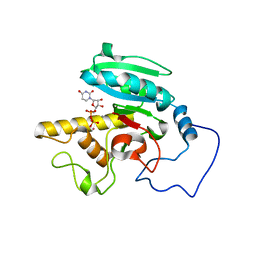 | | Glycosyltransferase B in complex with 3-amino-acceptor analog inhibitor and uridine diphosphate | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-galactosyltransferase, MANGANESE (II) ION, MERCURY (II) ION, ... | | Authors: | Nguyen, H.P, Seto, N.O.L, Cai, Y, Leinala, E.K, Borisova, S.N, Palcic, M.M, Evans, S.V. | | Deposit date: | 2003-10-22 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The influence of an intramolecular hydrogen bond in differential recognition of inhibitory acceptor analogs by human ABO(H) blood group A and B glycosyltransferases
J.Biol.Chem., 278, 2003
|
|
1R7V
 
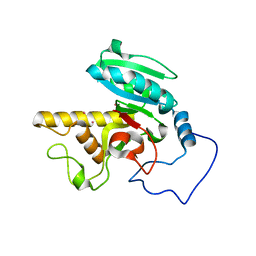 | | Glycosyltransferase A in complex with 3-amino-acceptor analog inhibitor | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-N-acetylgalactosaminyltransferase, MERCURY (II) ION, alpha-L-fucopyranose-(1-2)-hexyl 3-amino-3-deoxy-beta-D-galactopyranoside | | Authors: | Nguyen, H.P, Seto, N.O.L, Cai, Y, Leinala, E.K, Borisova, S.N, Palcic, M.M, Evans, S.V. | | Deposit date: | 2003-10-22 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The influence of an intramolecular hydrogen bond in differential recognition of inhibitory acceptor analogs by human ABO(H) blood group A and B glycosyltransferases
J.Biol.Chem., 278, 2003
|
|
1R7X
 
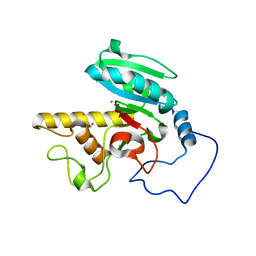 | | Glycosyltransferase B in complex with 3-amino-acceptor analog inhibitor | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-galactosyltransferase, MERCURY (II) ION, alpha-L-fucopyranose-(1-2)-hexyl 3-amino-3-deoxy-beta-D-galactopyranoside | | Authors: | Nguyen, H.P, Seto, N.O.L, Cai, Y, Leinala, E.K, Borisova, S.N, Palcic, M.M, Evans, S.V. | | Deposit date: | 2003-10-22 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The influence of an intramolecular hydrogen bond in differential recognition of inhibitory acceptor analogs by human ABO(H) blood group A and B glycosyltransferases
J.Biol.Chem., 278, 2003
|
|
1WT3
 
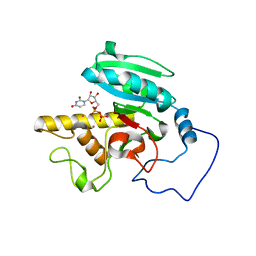 | | Mutant human ABO(H) blood group glycosyltransferase with bound UDP and acceptor | | Descriptor: | Histo-blood group ABO system transferase, MERCURY (II) ION, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Lee, H.J, Barry, C.H, Borisova, S.N, Seto, N.O.L, Zheng, R.B, Blancher, A, Evans, S.V, Palcic, M.M. | | Deposit date: | 2004-11-12 | | Release date: | 2004-12-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the inactivity of human blood group o2 glycosyltransferase
J.Biol.Chem., 280, 2005
|
|
3NE9
 
 | |
1R7Y
 
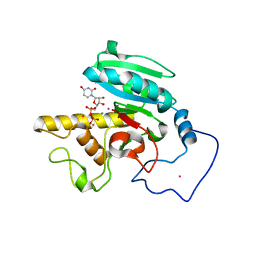 | | Glycosyltransferase A in complex with 3-amino-acceptor analog inhibitor and uridine diphosphate | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-N-acetylgalactosaminyltransferase, MANGANESE (II) ION, MERCURY (II) ION, ... | | Authors: | Nguyen, H.P, Seto, N.O.L, Cai, Y, Leinala, E.K, Borisova, S.N, Palcic, M.M, Evans, S.V. | | Deposit date: | 2003-10-22 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The influence of an intramolecular hydrogen bond in differential recognition of inhibitory acceptor analogs by human ABO(H) blood group A and B glycosyltransferases
J.Biol.Chem., 278, 2003
|
|
1R82
 
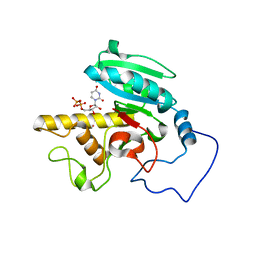 | | Glycosyltransferase B in complex with 3-amino-acceptor analog inhibitor, and uridine diphosphate-galactose | | Descriptor: | GALACTOSE-URIDINE-5'-DIPHOSPHATE, Glycoprotein-fucosylgalactoside alpha-galactosyltransferase, MERCURY (II) ION, ... | | Authors: | Nguyen, H.P, Seto, N.O.L, Cai, Y, Leinala, E.K, Borisova, S.N, Palcic, M.M, Evans, S.V. | | Deposit date: | 2003-10-22 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The influence of an intramolecular hydrogen bond in differential recognition of inhibitory acceptor analogs by human ABO(H) blood group A and B glycosyltransferases
J.Biol.Chem., 278, 2003
|
|
1VA5
 
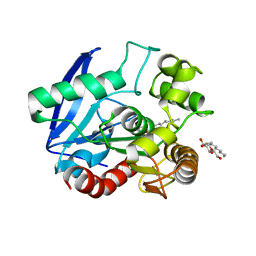 | | Antigen 85C with octylthioglucoside in active site | | Descriptor: | Antigen 85-C, octyl 1-thio-beta-D-glucopyranoside | | Authors: | Ronning, D.R, Vissa, V, Besra, G.S, Belisle, J.T, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-02-11 | | Release date: | 2004-07-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Mycobacterium tuberculosis Antigen 85A and 85C Structures Confirm Binding Orientation and Conserved Substrate Specificity
J.Biol.Chem., 279, 2004
|
|
1LZI
 
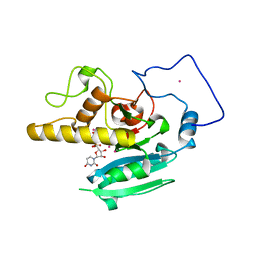 | | Glycosyltransferase A + UDP + H antigen acceptor | | Descriptor: | Glycosyltransferase A, MANGANESE (II) ION, MERCURY (II) ION, ... | | Authors: | Patenaude, S.I, Seto, N.O.L, Borisova, S.N, Szpacenko, A, Marcus, S.L, Palcic, M.M, Evans, S.V. | | Deposit date: | 2002-06-10 | | Release date: | 2002-08-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The structural basis for specificity
in human ABO(H) blood group biosynthesis.
Nat.Struct.Biol., 9, 2002
|
|
1J2Z
 
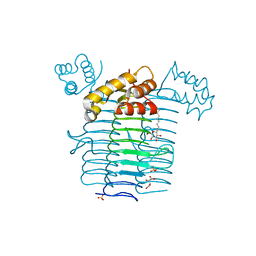 | | Crystal structure of UDP-N-acetylglucosamine acyltransferase | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, L(+)-TARTARIC ACID, SULFATE ION, ... | | Authors: | Lee, B.I, Suh, S.W. | | Deposit date: | 2003-01-15 | | Release date: | 2004-01-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of UDP-N-acetylglucosamine acyltransferase from Helicobacter pylori
Proteins, 53, 2003
|
|
1LZJ
 
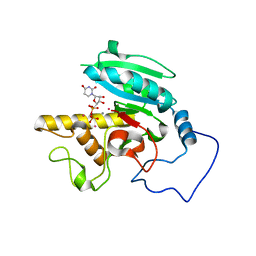 | | Glycosyltransferase B + UDP + H antigen acceptor | | Descriptor: | Glycosyltransferase B, MANGANESE (II) ION, MERCURY (II) ION, ... | | Authors: | Patenaude, S.I, Seto, N.O.L, Borisova, S.N, Szpacenko, A, Marcus, S.L, Palcic, M.M, Evans, S.V. | | Deposit date: | 2002-06-10 | | Release date: | 2002-08-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The structural basis for specificity in human
ABO(H) blood group biosynthesis.
Nat.Struct.Biol., 9, 2002
|
|
1C0O
 
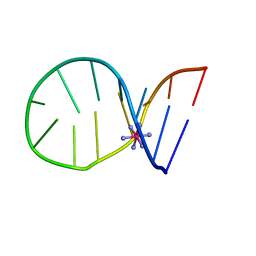 | |
2N89
 
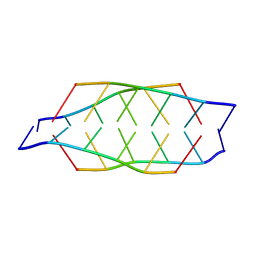 | | Tetrameric i-motif structure of dT-dC-dC-CFL-CFL-dC at acidic pH | | Descriptor: | DNA (5'-D(*TP*CP*CP*(CFL)P*(CFL)P*C)-3') | | Authors: | Abou-Assi, H, Harkness, R.W, Martin-Pintado, N, Wilds, C.J, Campos-Olivas, R, Mittermaier, A.K, Gonzalez, C, Damha, M.J. | | Deposit date: | 2015-10-09 | | Release date: | 2016-08-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Stabilization of i-motif structures by 2'-beta-fluorination of DNA.
Nucleic Acids Res., 44, 2016
|
|
1JKT
 
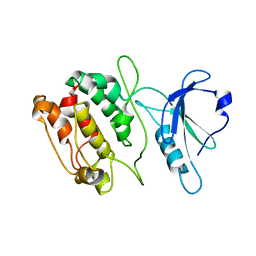 | | TETRAGONAL CRYSTAL FORM OF A CATALYTIC DOMAIN OF DEATH-ASSOCIATED PROTEIN KINASE | | Descriptor: | DEATH-ASSOCIATED PROTEIN KINASE | | Authors: | Tereshko, V, Teplova, M, Brunzelle, J, Watterson, D.M, Egli, M. | | Deposit date: | 2001-07-13 | | Release date: | 2002-04-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of the catalytic domain of human protein kinase associated with apoptosis and tumor suppression.
Nat.Struct.Biol., 8, 2001
|
|
2Y6K
 
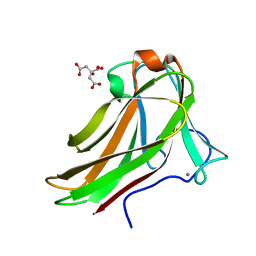 | | Xylotetraose bound to X-2 engineered mutated CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | Descriptor: | CALCIUM ION, CITRIC ACID, XYLANASE, ... | | Authors: | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2011-01-24 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
5L6Z
 
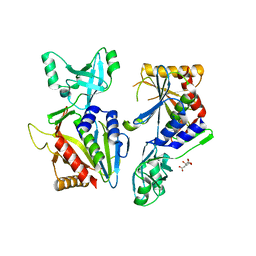 | |
4P6Y
 
 | |
6JV3
 
 | |
6Q5Q
 
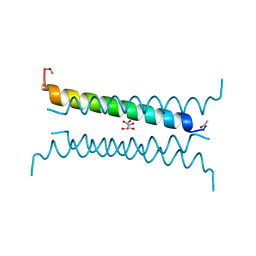 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex-KgEb | | Descriptor: | CC-Hex-KgEb, L(+)-TARTARIC ACID | | Authors: | Zaccai, N.R, Rhys, G.G, Wood, C.W, Beesley, J.L, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6Q5O
 
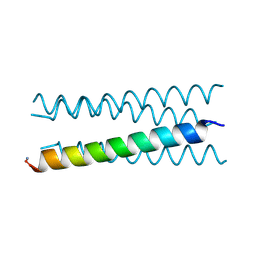 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex*-LL | | Descriptor: | CC-Hex*-LL | | Authors: | Rhys, G.G, Wood, C.W, Beesley, J.L, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6Q5R
 
 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex*-LL-KgEb | | Descriptor: | CC-Hex*-LL-KgEb, GLYCEROL | | Authors: | Beesley, J.L, Rhys, G.G, Wood, C.W, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6Q5H
 
 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex*-L24D | | Descriptor: | AMMONIUM ION, CC-Hex*-L24D, SULFATE ION | | Authors: | Wood, C.W, Beesley, J.L, Rhys, G.G, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6Q5K
 
 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex*-L24K | | Descriptor: | AMMONIUM ION, CC-Hex*-L24K, GLYCEROL, ... | | Authors: | Wood, C.W, Beesley, J.L, Rhys, G.G, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
