1MF2
 
 | | ANTI HIV1 PROTEASE FAB COMPLEX | | Descriptor: | MONOCLONAL ANTIBODY F11.2.32 | | Authors: | Lescar, J, Bentley, G.A. | | Deposit date: | 1996-12-27 | | Release date: | 1997-12-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional structure of an Fab-peptide complex: structural basis of HIV-1 protease inhibition by a monoclonal antibody.
J.Mol.Biol., 267, 1997
|
|
1MF4
 
 | | Structure-based design of potent and selective inhibitors of phospholipase A2: Crystal structure of the complex formed between phosholipase A2 from Naja Naja sagittifera and a designed peptide inhibitor at 1.9 A resolution | | Descriptor: | CALCIUM ION, Phospholipase A2, VAL-ALA-PHE-ARG-SER | | Authors: | Singh, R.K, Vikram, P, Paramsivam, M, Jabeen, T, Sharma, S, Makker, J, Dey, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2002-08-09 | | Release date: | 2003-09-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of specific peptide inhibitors for group I phospholipase A2: structure of a complex formed between phospholipase A2 from Naja naja sagittifera (group I) and a designed peptide inhibitor Val-Ala-Phe-Arg-Ser (VAFRS) at 1.9 A resolution reveals unique features
Biochemistry, 42, 2003
|
|
1MF5
 
 | | GCATGCT Quadruplex | | Descriptor: | 5'-D(*GP*CP*AP*TP*GP*CP*T)-3', COBALT HEXAMMINE(III) | | Authors: | Thorpe, J.H, Teixeira, S.C.M, Gale, B.C, Cardin, C.J. | | Deposit date: | 2002-08-09 | | Release date: | 2003-02-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of the complementary quadruplex formed by d(GCATGCT) at atomic
resolution
Nucleic Acids Res., 31, 2003
|
|
1MF6
 
 | |
1MF7
 
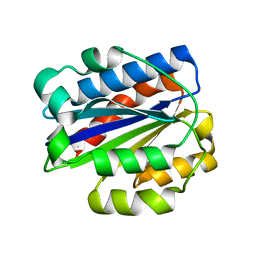 | | INTEGRIN ALPHA M I DOMAIN | | Descriptor: | INTEGRIN ALPHA M | | Authors: | McCleverty, C.J, Liddington, R.C. | | Deposit date: | 2002-08-09 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Engineered allosteric mutants of the integrin alphaMbeta2 I domain: structural and functional studies
Biochem.J., 372, 2003
|
|
1MF8
 
 | |
1MFA
 
 | |
1MFB
 
 | |
1MFC
 
 | |
1MFD
 
 | |
1MFE
 
 | |
1MFF
 
 | | MACROPHAGE MIGRATION INHIBITORY FACTOR Y95F MUTANT | | Descriptor: | MACROPHAGE MIGRATION INHIBITORY FACTOR | | Authors: | Taylor, A.B, Stamps, S.L, Wang, S.C, Hackert, M.L, Whitman, C.P. | | Deposit date: | 1998-10-19 | | Release date: | 1999-07-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of the phenylpyruvate tautomerase activity of macrophage migration inhibitory factor: properties of the P1G, P1A, Y95F, and N97A mutants.
Biochemistry, 39, 2000
|
|
1MFG
 
 | |
1MFI
 
 | | CRYSTAL STRUCTURE OF MACROPHAGE MIGRATION INHIBITORY FACTOR COMPLEXED WITH (E)-2-FLUORO-P-HYDROXYCINNAMATE | | Descriptor: | 2-FLUORO-3-(4-HYDROXYPHENYL)-2E-PROPENEOATE, PROTEIN (MACROPHAGE MIGRATION INHIBITORY FACTOR) | | Authors: | Taylor, A.B, Johnson Jr, W.H, Czerwinski, R.M, Whitman, C.P, Hackert, M.L. | | Deposit date: | 1998-08-12 | | Release date: | 1999-06-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of macrophage migration inhibitory factor complexed with (E)-2-fluoro-p-hydroxycinnamate at 1.8 A resolution: implications for enzymatic catalysis and inhibition.
Biochemistry, 38, 1999
|
|
1MFJ
 
 | |
1MFK
 
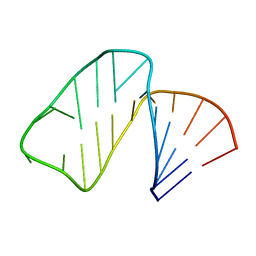 | | Structure of Prokaryotic SECIS mRNA Hairpin | | Descriptor: | 5'-R(P*GP*GP*CP*GP*GP*UP*UP*GP*CP*AP*GP*GP*UP*CP*UP*GP*CP*AP*CP*CP*GP*CP*C)-3' | | Authors: | Fourmy, D, Guittet, E, Yoshizawa, S. | | Deposit date: | 2002-08-12 | | Release date: | 2002-11-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of Prokaryotic SECIS mRNA Hairpin and its Interaction with Elongation Factor SELB
J.Mol.Biol., 324, 2002
|
|
1MFL
 
 | |
1MFM
 
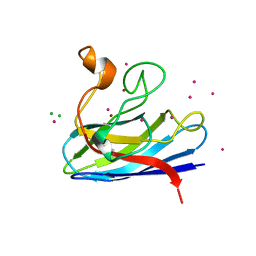 | | MONOMERIC HUMAN SOD MUTANT F50E/G51E/E133Q AT ATOMIC RESOLUTION | | Descriptor: | CADMIUM ION, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Ferraroni, M, Rypniewski, W, Wilson, K.S, Orioli, P.L, Viezzoli, M.S, Banci, L, Bertini, I, Mangani, S. | | Deposit date: | 1999-04-16 | | Release date: | 1999-04-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | The crystal structure of the monomeric human SOD mutant F50E/G51E/E133Q at atomic resolution. The enzyme mechanism revisited.
J.Mol.Biol., 288, 1999
|
|
1MFN
 
 | | SOLUTION NMR STRUCTURE OF LINKED CELL ATTACHMENT MODULES OF MOUSE FIBRONECTIN CONTAINING THE RGD AND SYNERGY REGIONS, 20 STRUCTURES | | Descriptor: | FIBRONECTIN | | Authors: | Copie, V, Tomita, Y, Akiyama, S.K, Aota, S, Yamada, K.M, Venable, R.M, Pastor, R.W, Krueger, S, Torchia, D.A. | | Deposit date: | 1998-01-27 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of linked cell attachment modules of mouse fibronectin containing the RGD and synergy regions: comparison with the human fibronectin crystal structure.
J.Mol.Biol., 277, 1998
|
|
1MFP
 
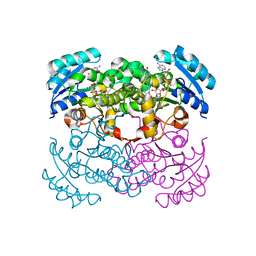 | | E. coli Enoyl Reductase in complex with NAD and SB611113 | | Descriptor: | (E)-N-METHYL-N-(1-METHYL-1H-INDOL-3-YLMETHYL)-3-(7-OXO-5,6,7,8-TETRAHYDRO-[1,8]NAPHTHYRIDIN-3-YL)-ACRYLAMIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Seefeld, M.A, Miller, W.H, Newlander, K.A, Burgess, W.J, DeWolf Jr, W.E, Elkins, P.A, Head, M.S, Jakas, D.R, Janson, C.A, Keller, P.M, Manley, P.J, Moore, T.D, Payne, D.J, Pearson, S, Polizzi, B.J, Qiu, X, Rittenhouse, S.F, Uzinskas, I.N, Wallis, N.G, Huffman, W.F. | | Deposit date: | 2002-08-13 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Indole Naphthyridinones as Inhibitors of Bacterial Enoyl-ACP Reductases FabI and FabK
J.MED.CHEM., 46, 2003
|
|
1MFQ
 
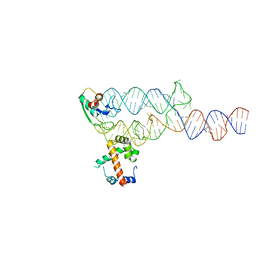 | | Crystal Structure Analysis of a Ternary S-Domain Complex of Human Signal Recognition Particle | | Descriptor: | 7S RNA of human SRP, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Kuglstatter, A, Oubridge, C, Nagai, K. | | Deposit date: | 2002-08-13 | | Release date: | 2002-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Induced structural changes of 7SL RNA during the assembly of human
signal recognition particle
Nat.Struct.Biol., 9, 2002
|
|
1MFR
 
 | | CRYSTAL STRUCTURE OF M FERRITIN | | Descriptor: | M FERRITIN, MAGNESIUM ION | | Authors: | Ha, Y, Shi, D, Allewell, N.M. | | Deposit date: | 1998-06-18 | | Release date: | 1999-06-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of bullfrog M ferritin at 2.8 A resolution: analysis of subunit interactions and the binuclear metal center
J.Biol.Inorg.Chem., 4, 1999
|
|
1MFS
 
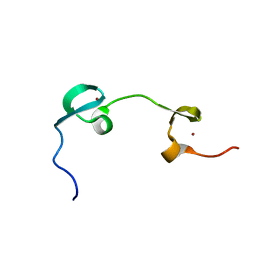 | | DYNAMICAL BEHAVIOR OF THE HIV-1 NUCLEOCAPSID PROTEIN; NMR, 30 STRUCTURES | | Descriptor: | HIV-1 NUCLEOCAPSID PROTEIN, ZINC ION | | Authors: | Summers, M.F, Turner, B.G, De Guzman, R.N, Lee, B.M, Tjandra, N. | | Deposit date: | 1998-04-01 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Dynamical behavior of the HIV-1 nucleocapsid protein.
J.Mol.Biol., 279, 1998
|
|
1MFT
 
 | | Crystal Structure Of Four-Helix Bundle Model | | Descriptor: | Four-helix bundle model, ZINC ION | | Authors: | Lahr, S.J, Stayrook, S.E, North, B, Kaplan, J, Geremia, S, DeGrado, W. | | Deposit date: | 2002-08-13 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Analysis and Design of Turns in alpha-Helical Hairpins
J.Mol.Biol., 346, 2005
|
|
1MFU
 
 | | Probing the role of a mobile loop in human salivary amylase: Structural studies on the loop-deleted mutant | | Descriptor: | 4-amino-4,6-dideoxy-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4-amino-4,6-dideoxy-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 5-HYDROXYMETHYL-CHONDURITOL, ... | | Authors: | Ramasubbu, N, Ragunath, C, Mishra, P.J. | | Deposit date: | 2002-08-13 | | Release date: | 2002-11-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the role of a mobile loop in substrate binding and enzyme activity of
human salivary amylase.
J.Mol.Biol., 325, 2003
|
|
