3MHL
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 4-{[N-(6-methoxy-5-nitropyrimidin-4-yl)amino]methyl}benzenesulfonamide | | Descriptor: | 4-{[(6-methoxy-5-nitropyrimidin-4-yl)amino]methyl}benzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Grazulis, S, Manakova, E, Golovenko, D. | | Deposit date: | 2010-04-08 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 4-[N-(Substituted 4-pyrimidinyl)amino]benzenesulfonamides as inhibitors of carbonic anhydrase isozymes I, II, VII, and XIII
Bioorg.Med.Chem., 18, 2010
|
|
3MYX
 
 | |
3KTJ
 
 | |
3KU4
 
 | | Trapping of an oxocarbenium ion intermediate in UP crystals | | Descriptor: | SULFATE ION, Uridine phosphorylase | | Authors: | Paul, D, O'Leary, S, Rajashankar, K, Bu, W, Toms, A, Settembre, E, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-11-26 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Glycal formation in crystals of uridine phosphorylase.
Biochemistry, 49, 2010
|
|
3KGS
 
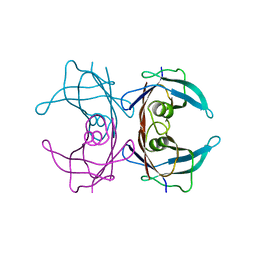 | | V30M mutant human transthyretin (TTR) (apoV30M) pH 7.5 | | Descriptor: | Transthyretin | | Authors: | Trivella, D.B, Polikarpov, I. | | Deposit date: | 2009-10-29 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational differences between the wild type and V30M mutant transthyretin modulate its binding to genistein: implications to tetramer stability and ligand-binding.
J.Struct.Biol., 170, 2010
|
|
3KGY
 
 | |
3KH9
 
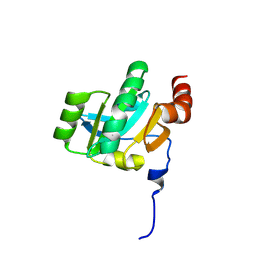 | | Crystal structure of the periplasmic soluble domain of oxidized CcmG from Pseudomonas aeruginosa | | Descriptor: | Thiol:disulfide interchange protein dsbE | | Authors: | Di Matteo, A, Calosci, N, Gianni, S, Jemth, P, Brunori, M, Travaglini Allocatelli, C. | | Deposit date: | 2009-10-30 | | Release date: | 2010-04-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional characterization of CcmG from Pseudomonas aeruginosa, a key component of the bacterial cytochrome c maturation apparatus.
Proteins, 78, 2010
|
|
3KJG
 
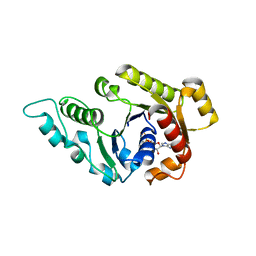 | | ADP-bound state of CooC1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CO dehydrogenase/acetyl-CoA synthase complex, accessory protein CooC | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2009-11-03 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the ATP-Dependent Maturation Factor of Ni,Fe-Containing Carbon Monoxide Dehydrogenases
J.Mol.Biol., 396, 2010
|
|
3KXF
 
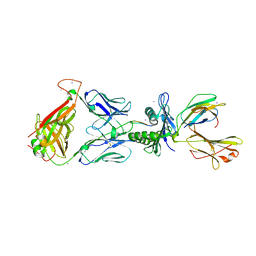 | | Crystal Structure of SB27 TCR in complex with the 'restriction triad' mutant HLA-B*3508-13mer | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-35 alpha chain, ... | | Authors: | Archbold, J.K, Tynan, F.E, Gras, S, Rossjohn, J. | | Deposit date: | 2009-12-03 | | Release date: | 2010-06-09 | | Last modified: | 2014-02-26 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Hard wiring of T cell receptor specificity for the major histocompatibility complex is underpinned by TCR adaptability
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KKF
 
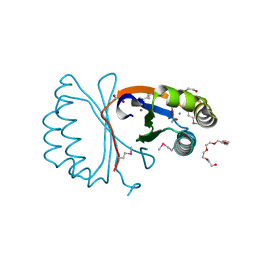 | |
3L0P
 
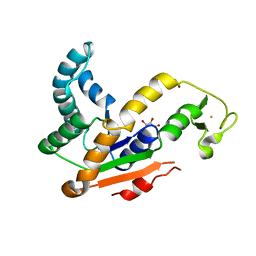 | | Crystal structures of Iron containing Adenylate kinase from Desulfovibrio gigas | | Descriptor: | Adenylate kinase, FE (III) ION, GLYCEROL | | Authors: | Mukhopadhyay, A, Trincao, J, Romao, M.J. | | Deposit date: | 2009-12-10 | | Release date: | 2010-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the zinc-, cobalt-, and iron-containing adenylate kinase from Desulfovibrio gigas: a novel metal-containing adenylate kinase from Gram-negative bacteria
J.Biol.Inorg.Chem., 16, 2011
|
|
3L16
 
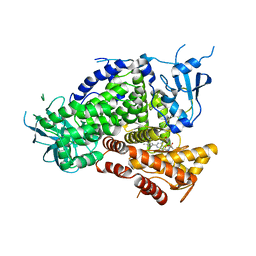 | | Discovery of (thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer | | Descriptor: | 5-(6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)pyridin-2-amine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Wiesmann, C. | | Deposit date: | 2009-12-10 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of (Thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer.
J.Med.Chem., 53, 2010
|
|
3KL9
 
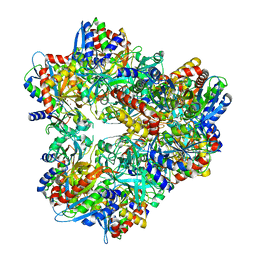 | | Crystal structure of PepA from Streptococcus pneumoniae | | Descriptor: | Glutamyl aminopeptidase, ZINC ION | | Authors: | Kim, K.K, Lee, S, Kim, D. | | Deposit date: | 2009-11-07 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the substrate specificity of PepA from Streptococcus pneumoniae, a dodecameric tetrahedral protease
Biochem.Biophys.Res.Commun., 391, 2010
|
|
3L1J
 
 | | Crystal structure of EstE5, was soaked by ZnSO4 | | Descriptor: | Esterase/lipase | | Authors: | Nam, K.H, Hwang, K.Y. | | Deposit date: | 2009-12-11 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the noninvasive inhibition of HSL-homolog EstE5 by organic solvents and metal ions
To be Published
|
|
3L1S
 
 | |
3L2W
 
 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with manganese and GS9137 (Elvitegravir) | | Descriptor: | 5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*TP*GP*TP*A)-3', 5'-D(*TP*AP*CP*AP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3', 6-(3-chloro-2-fluorobenzyl)-1-[(1S)-1-(hydroxymethyl)-2-methylpropyl]-7-methoxy-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, ... | | Authors: | Hare, S, Gupta, S.S, Cherepanov, P. | | Deposit date: | 2009-12-15 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Retroviral intasome assembly and inhibition of DNA strand transfer
Nature, 464, 2010
|
|
3KN8
 
 | |
3L3T
 
 | | Human mesotrypsin complexed with amyloid precursor protein inhibitor variant (APPIR15K) | | Descriptor: | CALCIUM ION, FORMIC ACID, PRSS3 protein, ... | | Authors: | Salameh, M.A, Soares, A.S, Radisky, E.S. | | Deposit date: | 2009-12-17 | | Release date: | 2010-09-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.378 Å) | | Cite: | Determinants of affinity and proteolytic stability in interactions of Kunitz family protease inhibitors with mesotrypsin.
J.Biol.Chem., 285, 2010
|
|
3KO4
 
 | |
3KOC
 
 | |
3L51
 
 | | Crystal Structure of the Mouse Condensin Hinge Domain | | Descriptor: | GLYCEROL, Structural maintenance of chromosomes protein 2, Structural maintenance of chromosomes protein 4 | | Authors: | Griese, J.J, Hopfner, K.-P. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.506 Å) | | Cite: | Structure and DNA binding activity of the mouse condensin hinge domain highlight common and diverse features of SMC proteins
Nucleic Acids Res., 38, 2010
|
|
3KEB
 
 | | Thiol peroxidase from Chromobacterium violaceum | | Descriptor: | CHLORIDE ION, Probable thiol peroxidase, SULFATE ION | | Authors: | Osipiuk, J, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-25 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of thiol peroxidase from Chromobacterium violaceum
To be Published
|
|
3KEP
 
 | | Crystal structure of the autoproteolytic domain from the nuclear pore complex component NUP145 from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, Nucleoporin NUP145 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Do, J, Bain, K, Dickey, M, Gheyi, T, Sali, A, Kim, S.J, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Rout, M, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145.
Proteins, 78, 2010
|
|
3KQV
 
 | | Crystal Structure of hPNMT in Complex AdoHcy and Formanilide | | Descriptor: | FORMANILIDE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
3KG9
 
 | |
