8UKQ
 
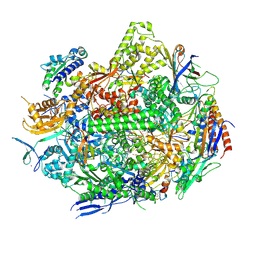 | | RNA polymerase II elongation complex with Fapy-dG lesion in apo state | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Hou, P, Oh, J, Wang, D. | | Deposit date: | 2023-10-15 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular Mechanism of RNA Polymerase II Transcriptional Mutagenesis by the Epimerizable DNA Lesion, Fapy·dG.
J.Am.Chem.Soc., 146, 2024
|
|
5UDQ
 
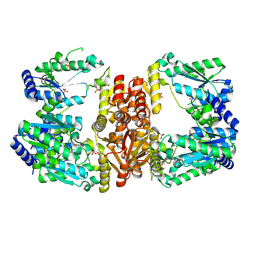 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase, apo form | | Descriptor: | Lactate racemization operon protein LarE, PHOSPHATE ION, SULFATE ION | | Authors: | Fellner, M, Desguin, B, Hausinger, R.P, Hu, J. | | Deposit date: | 2016-12-28 | | Release date: | 2017-08-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural insights into the catalytic mechanism of a sacrificial sulfur insertase of the N-type ATP pyrophosphatase family, LarE.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5CGD
 
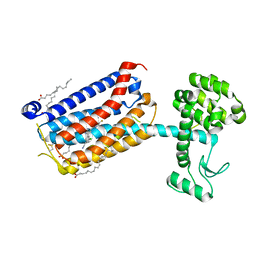 | | Structure of the human class C GPCR metabotropic glutamate receptor 5 transmembrane domain in complex with the negative allosteric modulator 3-chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile - (HTL14242) | | Descriptor: | 3-chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile, Metabotropic glutamate receptor 5,Endolysin,Metabotropic glutamate receptor 5, OLEIC ACID | | Authors: | Christopher, J.A, Aves, S.J, Bennett, K.A, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Okrasa, K, Serrano-Vega, M.J, Tehan, B.G, Wiggin, G.R, Congreve, M. | | Deposit date: | 2015-07-09 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Fragment and Structure-Based Drug Discovery for a Class C GPCR: Discovery of the mGlu5 Negative Allosteric Modulator HTL14242 (3-Chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile).
J.Med.Chem., 58, 2015
|
|
8RNY
 
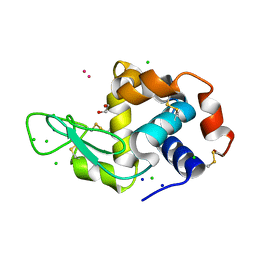 | | Hen Egg White Lysozyme soaked with with [H2Ind][trans-RuCl4(DMSO)(HInd)] | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Oszajca, M, Flejszar, M, Szura, A, Drozdz, P, Brindell, M, Kurpiewska, K. | | Deposit date: | 2024-01-11 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Exploring the coordination chemistry of ruthenium complexes with lysozymes: structural and in-solution studies.
Front Chem, 12, 2024
|
|
7OPV
 
 | | Crystal structure of the computationally designed SAKe6BE-3HH protein, alternative packing | | Descriptor: | SAKe6BE-3HH | | Authors: | Wouters, S.M.L, Noguchi, H, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-06-01 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
7OVU
 
 | |
8UKR
 
 | | RNA polymerase II elongation complex with Fapy-dG lesion soaking with ATP before chemistry | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Hou, P, Oh, J, Wang, D. | | Deposit date: | 2023-10-15 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.78 Å) | | Cite: | Molecular Mechanism of RNA Polymerase II Transcriptional Mutagenesis by the Epimerizable DNA Lesion, Fapy·dG.
J.Am.Chem.Soc., 146, 2024
|
|
5CKM
 
 | | The CUB1-EGF-CUB2 domains of rat MBL-associated serine protease-2 (MASP-2) bound to Ca2+ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Mannan-binding lectin serine peptidase 2 | | Authors: | Nan, R, Furze, C.M, Wright, D.W, Gor, J, Wallis, R, Perkins, S.J. | | Deposit date: | 2015-07-15 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Flexibility in Mannan-Binding Lectin-Associated Serine Proteases-1 and -2 Provides Insight on Lectin Pathway Activation.
Structure, 25, 2017
|
|
7OVV
 
 | | Crystal structure of the Arabidopsis thaliana thialysine acetyltransferase AtNATA2 | | Descriptor: | Probable acetyltransferase NATA1-like, [[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{R})-4-[[3-[2-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyldisulfanyl]ethylamino]-3-oxidanylidene-propyl]amino]-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Layer, D, Kopp, J, Sinning, I. | | Deposit date: | 2021-06-15 | | Release date: | 2022-12-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into the Arabidopsis thaliana thialysine acetyltransferase AtNATA2
To Be Published
|
|
5U0I
 
 | | C-terminal ankyrin repeats from human kidney-type glutaminase (KGA) - tetragonal crystal form | | Descriptor: | CHLORIDE ION, Glutaminase kidney isoform, mitochondrial, ... | | Authors: | Pasquali, C.C, Gonzalez, A, Dias, S.M.G, Ambrosio, A.L.B. | | Deposit date: | 2016-11-24 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.423 Å) | | Cite: | The origin and evolution of human glutaminases and their atypical C-terminal ankyrin repeats.
J. Biol. Chem., 292, 2017
|
|
8U8L
 
 | | X-ray crystal structure of TEBP-2 MCD3 with ds DNA | | Descriptor: | CESIUM ION, DNA (5'-D(*CP*TP*GP*TP*TP*AP*GP*GP*CP*TP*TP*AP*GP*GP*CP*TP*TP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*AP*AP*GP*CP*CP*TP*AP*AP*GP*CP*CP*TP*AP*AP*CP*A)-3'), ... | | Authors: | Nandakumar, J, Padmanaban, S. | | Deposit date: | 2023-09-18 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Caenorhabditis elegans telomere-binding proteins TEBP-1 and TEBP-2 adapt the Myb module to dimerize and bind telomeric DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7Z8O
 
 | | Crystal structure of SARS-CoV-2 S RBD in complex with a stapled peptide | | Descriptor: | 2,4,6-tris(chloromethyl)-1,3,5-triazine, GLYCEROL, Spike protein S1, ... | | Authors: | Brear, P, Chen, L, Gaynor, K, Harman, M, Dods, R, Hyvonen, M. | | Deposit date: | 2022-03-18 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Multivalent bicyclic peptides are an effective antiviral modality that can potently inhibit SARS-CoV-2.
Nat Commun, 14, 2023
|
|
7OWQ
 
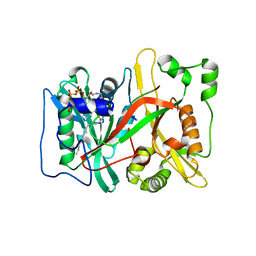 | |
7NB5
 
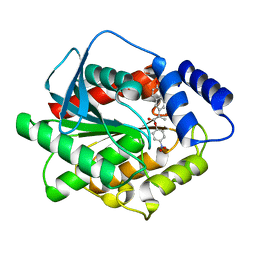 | |
7OWP
 
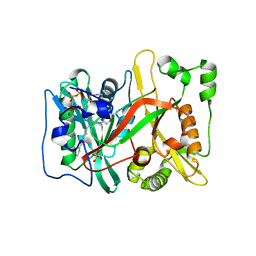 | | HsNMT1 in complex with both MyrCoA and ACE-G-(L-ORN)SFSKPR | | Descriptor: | ACE-GLY-ORN-SER-PHE-SER-LYS-PRO-ARG, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, ... | | Authors: | Dian, C, Giglione, C, Meinnel, T. | | Deposit date: | 2021-06-18 | | Release date: | 2022-12-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural and Large-scale Analysis Unveil the Intertwined Paths Promoting NMT-catalyzed Lysine and Glycine Myristoylation.
J.Mol.Biol., 434, 2022
|
|
5U0V
 
 | | E. coli dihydropteroate synthase complexed with 6-methylamino-5-nitrosoisocytosine | | Descriptor: | 2-amino-6-(methylamino)-5-nitrosopyrimidin-4(3H)-one, ACETIC ACID, Dihydropteroate synthase, ... | | Authors: | Peat, T.S, Dennis, M.L, Swarbrick, J.D. | | Deposit date: | 2016-11-27 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 8-Mercaptoguanine Derivatives as Inhibitors of Dihydropteroate Synthase.
Chemistry, 24, 2018
|
|
5GMA
 
 | |
8RV4
 
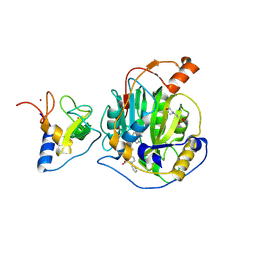 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 2 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-2-phenyl-benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
7NCF
 
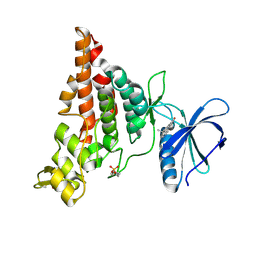 | | Crystal structure of HIPK2 in complex with MU135 (compound 21e) | | Descriptor: | 3-(4-Tert-butylphenyl)-5-(1H-pyrazol-4-yl)furo[3,2-b]pyridine, Homeodomain-interacting protein kinase 2 | | Authors: | Chaikuad, A, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-01-28 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Highly selective inhibitors of protein kinases CLK and HIPK with the furo[3,2-b]pyridine core.
Eur.J.Med.Chem., 215, 2021
|
|
7P4O
 
 | | Crystal structure of Autotaxin and 9(R)-delta6a,10a-THC | | Descriptor: | (9~{R},10~{a}~{S})-6,6,9-trimethyl-3-pentyl-6~{a},7,8,9,10,10~{a}-hexahydrobenzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7alpha-hydroxycholesterol, ... | | Authors: | Eymery, M.C, McCarthy, A.A, Hausmann, J. | | Deposit date: | 2021-07-12 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Linking medicinal cannabis to autotaxin-lysophosphatidic acid signaling.
Life Sci Alliance, 6, 2023
|
|
6MKG
 
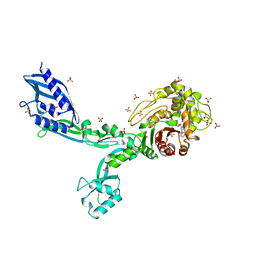 | | Crystal structure of penicillin binding protein 5 (PBP5) from Enterococcus faecium in the benzylpenicilin-bound form | | Descriptor: | OPEN FORM - PENICILLIN G, SULFATE ION, penicillin binding protein 5 (PBP5) | | Authors: | Moon, T.M, Lee, C, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
7OWU
 
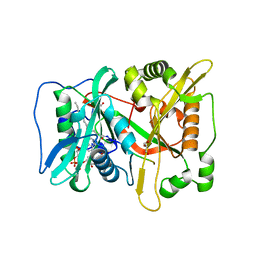 | | HsNMT1 in complex with both CoA and Myr-ANCFSKPR peptide | | Descriptor: | ALA-ASN-CYS-PHE-SER-LYS-PRO-ARG, COENZYME A, GLYCEROL, ... | | Authors: | Dian, C, Giglione, C, Meinnel, T. | | Deposit date: | 2021-06-18 | | Release date: | 2022-12-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural and Large-scale Analysis Unveil the Intertwined Paths Promoting NMT-catalyzed Lysine and Glycine Myristoylation.
J.Mol.Biol., 434, 2022
|
|
7V6M
 
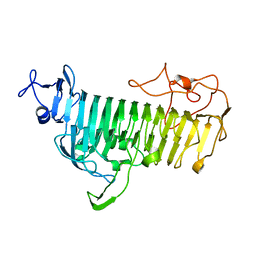 | |
5GM1
 
 | | Crystal structure of methyltransferase TleD complexed with SAH | | Descriptor: | O-methylransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Yu, F, Li, M.J, Xu, C.Y, Zhou, H, Sun, B, Wang, Z.J, Xu, Q, Xie, M.Y, Zuo, G, Huang, P, Wang, Q.S, He, J.H. | | Deposit date: | 2016-07-12 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal structure and enantioselectivity of terpene cyclization in SAM-dependent methyltransferase TleD
Biochem.J., 473, 2016
|
|
7OWO
 
 | | HsNMT1 in complex with both MyrCoA and N-acetylated KSFSKPR peptide | | Descriptor: | COENZYME A, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, ... | | Authors: | Dian, C, Giglione, C, Meinnel, T. | | Deposit date: | 2021-06-18 | | Release date: | 2022-12-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Large-scale Analysis Unveil the Intertwined Paths Promoting NMT-catalyzed Lysine and Glycine Myristoylation.
J.Mol.Biol., 434, 2022
|
|
