8UDV
 
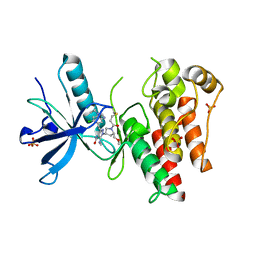 | | The X-RAY co-crystal structure of human FGFR3 V555M and Compound 17 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(6-chloro-1-cyclopropyl-1H-benzimidazol-5-yl)ethynyl]-1-[(3S,5S)-5-(methoxymethyl)-1-(prop-2-enoyl)pyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, Fibroblast growth factor receptor 3, ... | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
5C9S
 
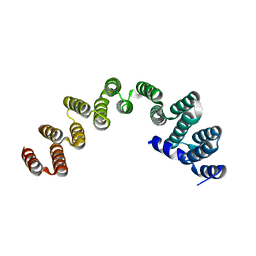 | |
8KCK
 
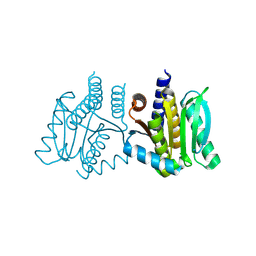 | | De novo design protein -N9 | | Descriptor: | De novo design protein -N9 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-07 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | De novo design protein -N9
To Be Published
|
|
7MU6
 
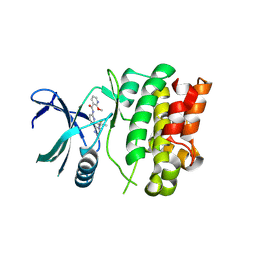 | | Ask1 bound to compound 28 | | Descriptor: | 2-methoxy-N-(6-{4-[(2S)-1,1,1-trifluoropropan-2-yl]-4H-1,2,4-triazol-3-yl}pyridin-2-yl)pyridine-3-carboxamide, Mitogen-activated protein kinase kinase kinase 5 | | Authors: | Chodaparambil, J.V, Marcotte, D.J. | | Deposit date: | 2021-05-14 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.165 Å) | | Cite: | Discovery of Potent, Selective, and Brain-Penetrant Apoptosis Signal-Regulating Kinase 1 (ASK1) Inhibitors that Modulate Brain Inflammation In Vivo.
J.Med.Chem., 64, 2021
|
|
6N0M
 
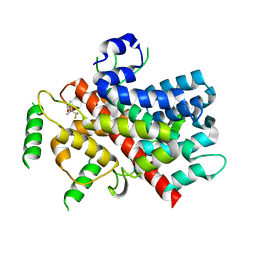 | |
7NFU
 
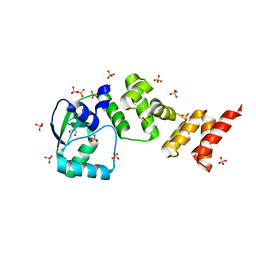 | | Crystal structure of C-terminally truncated Geobacillus thermoleovorans nucleoid occlusion protein Noc | | Descriptor: | GLYCEROL, Nucleoid occlusion protein, SULFATE ION | | Authors: | Jalal, A.S.B, Tran, N.T, Wu, L.J, Ramakrishnan, K, Rejzek, M, Stevenson, C.E.M, Lawson, D.M, Errington, J, Le, T.B.K. | | Deposit date: | 2021-02-07 | | Release date: | 2021-02-17 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | CTP regulates membrane-binding activity of the nucleoid occlusion protein Noc.
Mol.Cell, 81, 2021
|
|
8U5M
 
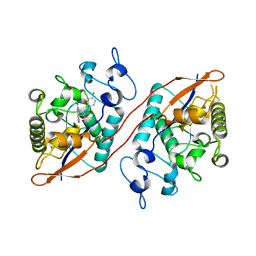 | | Structure of Sts-1 HP domain with rebamipide | | Descriptor: | Rebamipide, Ubiquitin-associated and SH3 domain-containing protein B | | Authors: | Azia, F, Dey, R, French, J.B. | | Deposit date: | 2023-09-12 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Rebamipide and Derivatives are Potent, Selective Inhibitors of Histidine Phosphatase Activity of the Suppressor of T Cell Receptor Signaling Proteins.
J.Med.Chem., 67, 2024
|
|
7NFW
 
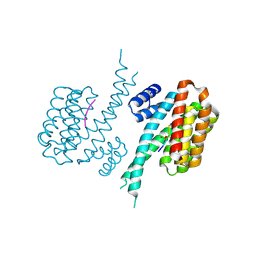 | |
6N1B
 
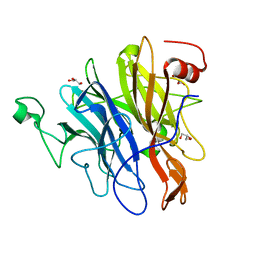 | | Crystal structure of an N-acetylgalactosamine deacetylase from F. plautii in complex with blood group B trisaccharide | | Descriptor: | CALCIUM ION, Carbohydrate-binding protein, GLYCEROL, ... | | Authors: | Sim, L, Rahfeld, P, Withers, S.G. | | Deposit date: | 2018-11-08 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An enzymatic pathway in the human gut microbiome that converts A to universal O type blood.
Nat Microbiol, 4, 2019
|
|
7OQV
 
 | |
7OQ9
 
 | |
7NG0
 
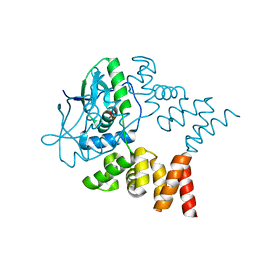 | | Crystal structure of N- and C-terminally truncated Geobacillus thermoleovorans nucleoid occlusion protein Noc | | Descriptor: | Nucleoid occlusion protein, SULFATE ION | | Authors: | Jalal, A.S.B, Tran, N.T, Wu, L.J, Ramakrishnan, K, Rejzek, M, Stevenson, C.E.M, Lawson, D.M, Errington, J, Le, T.B.K. | | Deposit date: | 2021-02-08 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | CTP regulates membrane-binding activity of the nucleoid occlusion protein Noc.
Mol.Cell, 81, 2021
|
|
7PA2
 
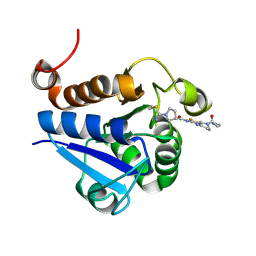 | | PARK7 with inhibitor 8RK64 | | Descriptor: | (3~{S})-~{N}-[5-[2-[(azanylidene-$l^{4}-azanylidene)amino]ethanoyl]-6,7-dihydro-4~{H}-[1,3]thiazolo[5,4-c]pyridin-2-yl]-1-(iminomethyl)pyrrolidine-3-carboxamide, Parkinson disease protein 7 | | Authors: | Kim, R.Q, Jia, Y, Sapmaz, A, Geurink, P.P. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Chemical Toolkit for PARK7: Potent, Selective, and High-Throughput.
J.Med.Chem., 65, 2022
|
|
7PA3
 
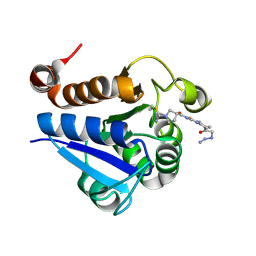 | | PARK7 with covalent inhibitor JYQ-88 | | Descriptor: | (3~{S})-~{N}-[5-[2-[(azanylidene-$l^{4}-azanylidene)amino]ethanoyl]-6,7-dihydro-4~{H}-[1,3]thiazolo[5,4-c]pyridin-2-yl]-1-(iminomethyl)pyrrolidine-3-carboxamide, Parkinson disease protein 7 | | Authors: | Kim, R.Q, Jia, Y, Sapmaz, A, Geurink, P.P. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Chemical Toolkit for PARK7: Potent, Selective, and High-Throughput.
J.Med.Chem., 65, 2022
|
|
5GGK
 
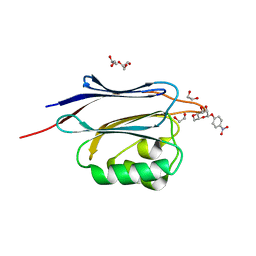 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with Man-beta-pNP | | Descriptor: | 1,2-ETHANEDIOL, 4-nitrophenyl beta-D-mannopyranoside, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7OQA
 
 | |
5GGO
 
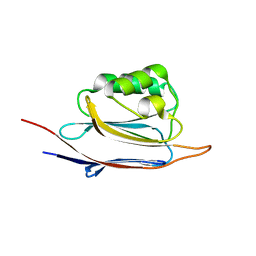 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with GalNac-beta1,3-GlcNAc-beta-pNP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-4-nitrophenyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1 | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
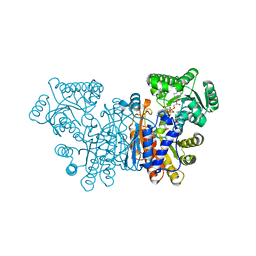
7NED
 
 | |
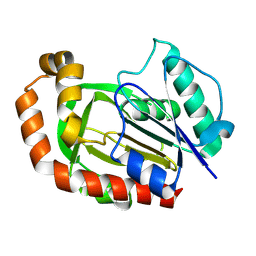
8KHS
 
 | |
7ONG
 
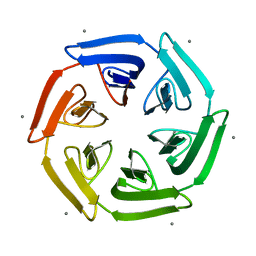 | | Crystal structure of the computationally designed SAKe6BE-L1 protein | | Descriptor: | CALCIUM ION, SAKe6BE-L1 | | Authors: | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-05-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
7OPU
 
 | |
5U9W
 
 | | Structure of CNTnw N149L in the intermediate 3 state | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-beta-D-glucopyranosyl)oxy]methyl}-2-octyldecyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, Nucleoside permease | | Authors: | Hirschi, M, Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2016-12-18 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.555 Å) | | Cite: | Visualizing multistep elevator-like transitions of a nucleoside transporter.
Nature, 545, 2017
|
|
6N31
 
 | | WD repeats of human WDR12 | | Descriptor: | Ribosome biogenesis protein WDR12, UNKNOWN ATOM OR ION | | Authors: | Halabelian, L, Zeng, H, Tempel, W, Li, Y, Seitova, A, Hutchinson, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-14 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | WD repeats of human WDR12
To Be Published
|
|
7ON7
 
 | |
7OL2
 
 | | Crystal structure of mouse contactin 1 immunoglobulin domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin-1, ... | | Authors: | Chataigner, L.M.P, Janssen, B.J.C. | | Deposit date: | 2021-05-19 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.89 Å) | | Cite: | Structural insights into the contactin 1 - neurofascin 155 adhesion complex.
Nat Commun, 13, 2022
|
|
