8KBL
 
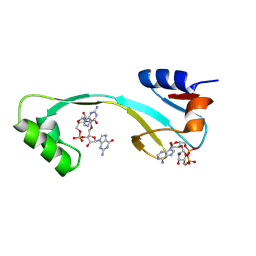 | | Structure of AcrIIA7 complexed with 1',3'-cADPR and cGG | | Descriptor: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Inhibitor of Type II CRISPR-Cas system | | Authors: | Xiao, Y, Feng, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of AcrIIA7 complexed with 1',3'-cADPR and cGG
To Be Published
|
|
5CDF
 
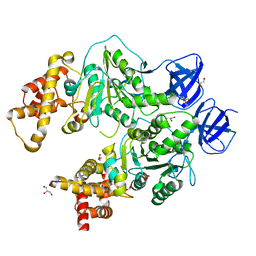 | | Structure at 2.3 A of the alpha/beta monomer of the F-ATPase from Paracoccus denitrificans | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, GLYCEROL, ... | | Authors: | Morales-Rios, E, Montgomery, M.G, Leslie, A.G.W, Garcia-Trejo, J.J, Walker, J.E. | | Deposit date: | 2015-07-03 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a catalytic dimer of the alpha- and beta-subunits of the F-ATPase from Paracoccus denitrificans at 2.3 angstrom resolution.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
5CDQ
 
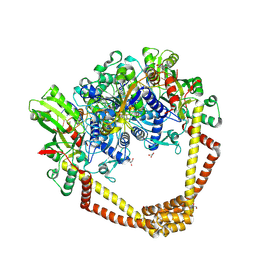 | | 2.95A structure of Moxifloxacin with S.aureus DNA gyrase and DNA | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP*T*GP*GP*CP*CP*AP*TP*AP*CP*GP*CP*TP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5TQN
 
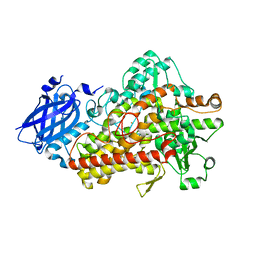 | | Lipoxygenase-1 (soybean) L546A mutant at 293K | | Descriptor: | FE (II) ION, Seed linoleate 13S-lipoxygenase-1 | | Authors: | Poss, E.M, Fraser, J.S, Gee, C. | | Deposit date: | 2016-10-24 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biophysical Characterization of a Disabled Double Mutant of Soybean Lipoxygenase: The "Undoing" of Precise Substrate Positioning Relative to Metal Cofactor and an Identified Dynamical Network.
J.Am.Chem.Soc., 141, 2019
|
|
8KBD
 
 | |
8KC0
 
 | | De novo design protein -NB8 | | Descriptor: | De novo design protein -NB8 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-05 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | De novo design protein -NB8
To Be Published
|
|
7P8O
 
 | | Crystal structure of D-aminoacid transaminase from Haliscomenobacter hydrossis in its intermediate form | | Descriptor: | Aminotransferase class IV, MAGNESIUM ION, SULFATE ION | | Authors: | Matyuta, I.O, Boyko, K.M, Bakunova, A.K, Nikolaeva, A.Y, Rakitina, T.V, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2021-07-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Incorporation of pyridoxal-5'-phosphate into the apoenzyme: A structural study of D-amino acid transaminase from Haliscomenobacter hydrossis.
Biochim Biophys Acta Proteins Proteom, 1873, 2024
|
|
5CEF
 
 | |
8KBJ
 
 | | Structure of HgmTad2 complexed with 1',2'-cADPR | | Descriptor: | (1S,3R,4R,6R,9S,11R,14R,15S,16R,18R)-4-(6-amino-9H-purin-9-yl)-9,11,15,16,18-pentahydroxy-2,5,8,10,12,17-hexaoxa-9lambda~5~,11lambda~5~-diphosphatricyclo[12.2.1.1~3,6~]octadecane-9,11-dione, Inhibitor of Type II CRISPR-Cas system | | Authors: | Xiao, Y, Feng, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of HgmTad2 complexed with 1',2'-cADPR
To Be Published
|
|
5U5K
 
 | |
8KBG
 
 | |
8V3K
 
 | | F95S-F198S epi-Isozizaene Synthase: complex with 3 Mg2+, inorganic pyrophosphate, and benzyl triethyl ammonium cation | | Descriptor: | DIPHOSPHATE, Epi-isozizaene synthase, GLYCEROL, ... | | Authors: | Christianson, D.W, Eaton, S.A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure-Based Engineering of a Sesquiterpene Cyclase to Generate an Alcohol Product: Conversion of epi -Isozizaene Synthase into alpha-Bisabolol Synthase.
Biochemistry, 63, 2024
|
|
8KBC
 
 | | Structure of CmTad1 complexed with cAAA | | Descriptor: | (2-ACETYL-5-METHYLANILINO)(2,6-DIBROMOPHENYL)ACETAMIDE, Thoeris anti-defense 1, ZINC ION | | Authors: | Xiao, Y, Feng, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of CmTad1 complexed with cAAA
To Be Published
|
|
7P8J
 
 | | Receptor-binding domain (RBD) of the spike protein of the bat coronavirus RaTG13 virus in complex with the extracellular domain of human angiotensin-converting enzyme 2 (ACE2) - Crystal form 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Scietti, L, Castelli, M, Faravelli, S, Clementi, N, Mancini, N, Forneris, F. | | Deposit date: | 2021-07-22 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (6.585 Å) | | Cite: | Constrained Evolution of SARS-CoV-2 Spike in Rhinolophus affinis Bats
To Be Published
|
|
5H2V
 
 | |
6MRY
 
 | | NoD173 plant defensin | | Descriptor: | 1,2-ETHANEDIOL, 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, CHLORIDE ION, ... | | Authors: | Caria, S, Kvansakul, M. | | Deposit date: | 2018-10-15 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional characterization of the membrane-permeabilizing activity ofNicotiana occidentalisdefensin NoD173 and protein engineering to enhance oncolysis.
Faseb J., 33, 2019
|
|
8KFI
 
 | |
6MXR
 
 | |
7P8I
 
 | | Receptor-binding domain (RBD) of the spike protein of the bat coronavirus RaTG13 virus in complex with the extracellular domain of human angiotensin-converting enzyme 2 (ACE2) - Crystal form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Scietti, L, Castelli, M, Faravelli, S, Clementi, N, Mancini, N, Forneris, F. | | Deposit date: | 2021-07-22 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Evidence of SARS-CoV-2 Direct Evolution in R. affinis Bats Driven by Affinity and Dynamics Optimization of the Spike Protein
To Be Published
|
|
8SBM
 
 | | Crystal structure of the wild-type Catalytic ATP-binding domain of Mtb DosS | | Descriptor: | 1,2-ETHANEDIOL, GAF domain-containing protein, SODIUM ION, ... | | Authors: | Larson, G, Shi, K, Aihara, H, Bhagi-Damodaran, A. | | Deposit date: | 2023-04-03 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Understanding ATP Binding to DosS Catalytic Domain with a Short ATP-Lid.
Biochemistry, 62, 2023
|
|
5CFO
 
 | | Crystal structure of anemone STING (Nematostella vectensis) in apo 'rotated' open conformation | | Descriptor: | Stimulator of Interferon Genes | | Authors: | Kranzusch, P.J, Wilson, S.C, Lee, A.S.Y, Berger, J.M, Doudna, J.A, Vance, R.E. | | Deposit date: | 2015-07-08 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2',3' cGAMP Signaling.
Mol.Cell, 59, 2015
|
|
8SBS
 
 | |
5H4G
 
 | | Structure of PIN-domain protein (VapC4 toxin) from Pyrococcus horikoshii determined at 1.77 A resolution | | Descriptor: | Ribonuclease VapC4, ZINC ION | | Authors: | Biswas, A, Hatti, K, Srinivasan, N, Murthy, M.R.N, Sekar, K. | | Deposit date: | 2016-10-31 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure determination of contaminant proteins using the MarathonMR procedure
J. Struct. Biol., 197, 2017
|
|
7P9W
 
 | | Epstein-Barr virus encoded apoptosis regulator BHRF1 in complex with Puma BH3 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, AMMONIUM ION, ... | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.00010061 Å) | | Cite: | Crystal Structures of Epstein-Barr Virus Bcl-2 Homolog BHRF1 Bound to Bid and Puma BH3 Motif Peptides.
Viruses, 14, 2022
|
|
8KFQ
 
 | |
