2K1E
 
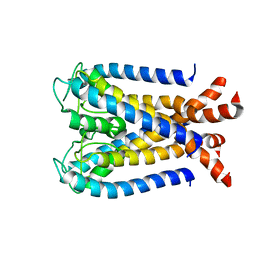 | | NMR studies of a channel protein without membranes: structure and dynamics of water-solubilized KcsA | | Descriptor: | water soluble analogue of potassium channel, KcsA | | Authors: | Ma, D, Xu, Y, Tillman, T, Tang, P, Meirovitch, E, Eckenhoff, R, Carnini, A. | | Deposit date: | 2008-02-29 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR studies of a channel protein without membranes: structure and dynamics of water-solubilized KcsA.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3LQ6
 
 | |
3ONC
 
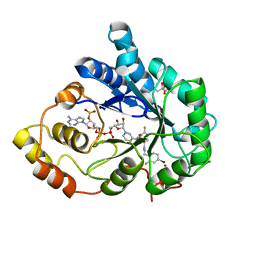 | | Bond breakage and relocation of a covalently bound bromine of IDD594 in a complex with hAR T113A mutant after moderate radiation dose | | Descriptor: | Aldose reductase, BROMIDE ION, CITRIC ACID, ... | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-08-28 | | Release date: | 2011-08-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Radiation damage reveals promising interaction position
J.SYNCHROTRON RADIAT., 18, 2011
|
|
2L5K
 
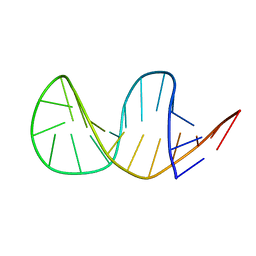 | | Solution structure of truncated 23-mer DNA MUC1 aptamer | | Descriptor: | DNA (5'-R(*(N68)P*G)-D(*CP*AP*GP*TP*TP*GP*AP*TP*CP*CP*TP*TP*TP*GP*GP*AP*TP*AP*CP*CP*CP*TP*GP*GP*T)-3') | | Authors: | Cognet, J, Baouendi, M, Hantz, E, Missailidis, S, Herve du Penhoat, C, Piotto, M. | | Deposit date: | 2010-11-02 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a truncated anti-MUC1 DNA aptamer determined by mesoscale modeling and NMR.
Febs J., 279, 2012
|
|
2LGI
 
 | | Atomic Resolution Protein Structures using NMR Chemical Shift Tensors | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Wylie, B.J, Sperling, L.J, Nieuwkoop, A.J, Franks, W.T, Oldfield, E, Rienstra, C.M. | | Deposit date: | 2011-07-26 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Ultrahigh resolution protein structures using NMR chemical shift tensors.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OJ7
 
 | |
4DXY
 
 | | Crystal structures of CYP101D2 Y96A mutant | | Descriptor: | Cytochrome P450, DI(HYDROXYETHYL)ETHER, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhou, W, Bell, S.G, Yang, W, Dale, A, Wong, L.-L. | | Deposit date: | 2012-02-28 | | Release date: | 2012-08-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Improving the affinity and activity of CYP101D2 for hydrophobic substrates
Appl.Microbiol.Biotechnol., 2012
|
|
1X42
 
 | | Crystal structure of a haloacid dehalogenase family protein (PH0459) from Pyrococcus horikoshii OT3 | | Descriptor: | hypothetical protein PH0459 | | Authors: | Arai, R, Kukimoto-Niino, M, Sugahara, M, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-12 | | Release date: | 2005-11-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the probable haloacid dehalogenase PH0459 from Pyrococcus horikoshii OT3
Protein Sci., 15, 2006
|
|
2J5I
 
 | | Crystal Structure of Hydroxycinnamoyl-CoA Hydratase-Lyase | | Descriptor: | P-HYDROXYCINNAMOYL COA HYDRATASE/LYASE | | Authors: | Leonard, P.M, Brzozowski, A.M, Lebedev, A, Marshall, C.M, Smith, D.J, Verma, C.S, Walton, N.J, Grogan, G. | | Deposit date: | 2006-09-18 | | Release date: | 2006-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8 A Resolution Structure of Hydroxycinnamoyl- Coenzyme a Hydratase-Lyase (Hchl) from Pseudomonas Fluorescens, an Enzyme that Catalyses the Transformation of Feruloyl-Coenzyme a to Vanillin.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2RAR
 
 | |
2RAV
 
 | |
2D2E
 
 | | Crystal structure of atypical cytoplasmic ABC-ATPase SufC from Thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, GLYCEROL, SufC protein | | Authors: | Watanabe, S, Kita, A, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-09-08 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Atypical Cytoplasmic ABC-ATPase SufC from Thermus thermophilus HB8.
J.Mol.Biol., 353, 2005
|
|
2RBK
 
 | |
3EHA
 
 | |
3VCY
 
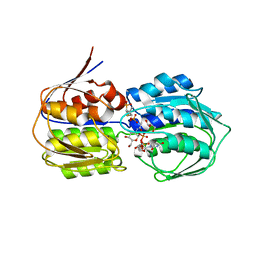 | | Structure of MurA (UDP-N-acetylglucosamine enolpyruvyl transferase), from Vibrio fischeri in complex with substrate UDP-N-acetylglucosamine and the drug fosfomycin. | | Descriptor: | GLYCEROL, PHOSPHATE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, ... | | Authors: | Bensen, D.C, Rodriguez, S, Nix, J, Cunningham, M.L, Tari, L.W. | | Deposit date: | 2012-01-04 | | Release date: | 2012-04-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | Structure of MurA (UDP-N-acetylglucosamine enolpyruvyl transferase) from Vibrio fischeri in complex with substrate UDP-N-acetylglucosamine and the drug fosfomycin.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2NC7
 
 | |
2Z26
 
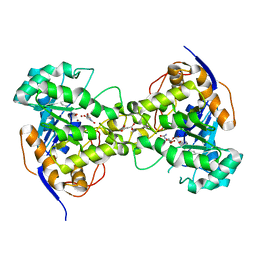 | | Thr110Ala dihydroorotase from E. coli | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, MALONATE ION, ... | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2007-05-17 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Kinetic and Structural Analysis of Mutant Escherichia coli Dihydroorotases: A Flexible Loop Stabilizes the Transition State
Biochemistry, 46, 2007
|
|
2Z28
 
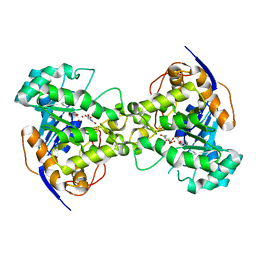 | | Thr109Val dihydroorotase from E. coli | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ... | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2007-05-17 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Kinetic and Structural Analysis of Mutant Escherichia coli Dihydroorotases: A Flexible Loop Stabilizes the Transition State
Biochemistry, 46, 2007
|
|
2Z24
 
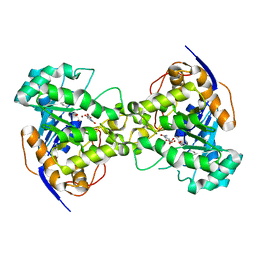 | | Thr110Ser dihydroorotase from E. coli | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ... | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2007-05-17 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and Structural Analysis of Mutant Escherichia coli Dihydroorotases: A Flexible Loop Stabilizes the Transition State
Biochemistry, 46, 2007
|
|
2Z2B
 
 | |
3LQY
 
 | | Crystal structure of putative isochorismatase hydrolase from Oleispira antarctica | | Descriptor: | GLYCEROL, putative isochorismatase hydrolase | | Authors: | Goral, A, Chruszcz, M, Kagan, O, Cymborowski, M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-10 | | Release date: | 2010-03-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a putative isochorismatase hydrolase from Oleispira antarctica.
J.Struct.Funct.Genom., 13, 2012
|
|
2Z25
 
 | | Thr110Val dihydroorotase from E. coli | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ... | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2007-05-17 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Kinetic and Structural Analysis of Mutant Escherichia coli Dihydroorotases: A Flexible Loop Stabilizes the Transition State
Biochemistry, 46, 2007
|
|
3PRC
 
 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS (QB-DEPLETED) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Lancaster, C.R.D, Michel, H. | | Deposit date: | 1997-07-29 | | Release date: | 1998-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The coupling of light-induced electron transfer and proton uptake as derived from crystal structures of reaction centres from Rhodopseudomonas viridis modified at the binding site of the secondary quinone, QB.
Structure, 5, 1997
|
|
2NM3
 
 | | Crystal structure of dihydroneopterin aldolase from S. aureus in complex with (1S,2S)-monapterin at 1.68 angstrom resolution | | Descriptor: | ACETATE ION, D-MONAPTERIN, Dihydroneopterin aldolase | | Authors: | Blaszczyk, J, Ji, X, Yan, H. | | Deposit date: | 2006-10-20 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis for the aldolase and epimerase activities of Staphylococcus aureus dihydroneopterin aldolase.
J.Mol.Biol., 368, 2007
|
|
3CZA
 
 | | Crystal Structure of E18D DJ-1 | | Descriptor: | MALONIC ACID, Protein DJ-1 | | Authors: | Witt, A.C, Lakshminarasimhan, M, Remington, B.C, Hashim, S, Pozharski, E, Wilson, M.A. | | Deposit date: | 2008-04-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Cysteine pKa depression by a protonated glutamic acid in human DJ-1.
Biochemistry, 47, 2008
|
|
