3LEW
 
 | |
3LVL
 
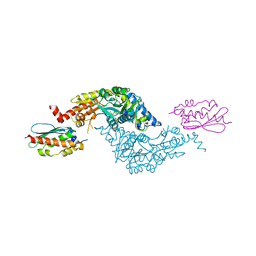 | | Crystal Structure of E.coli IscS-IscU complex | | Descriptor: | Cysteine desulfurase, NifU-like protein, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Shi, R, Proteau, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-02-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions.
Plos Biol., 8, 2010
|
|
3LWC
 
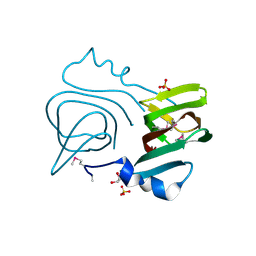 | |
3LH1
 
 | | Q191A mutant of the DegS-deltaPDZ | | Descriptor: | Protease degS | | Authors: | Sohn, J, Grant, R.A, Sauer, R.T. | | Deposit date: | 2010-01-21 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.507 Å) | | Cite: | Allostery is an intrinsic property of the protease domain of DegS: implications for enzyme function and evolution.
J.Biol.Chem., 285, 2010
|
|
3LHQ
 
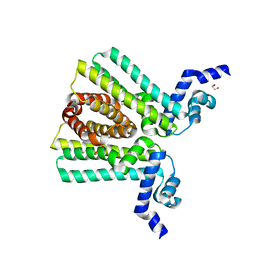 | | DNA-binding transcriptional repressor AcrR from Salmonella typhimurium. | | Descriptor: | 1,2-ETHANEDIOL, AcrAB operon repressor (TetR/AcrR family), DI(HYDROXYETHYL)ETHER | | Authors: | Osipiuk, J, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-22 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | X-ray crystal structure of DNA-binding transcriptional repressor AcrR from Salmonella typhimurium.
To be Published
|
|
3LX5
 
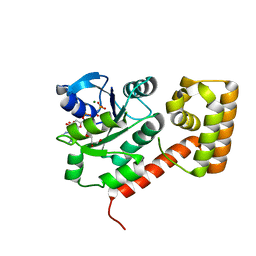 | | Crystal structure of mGMPPNP-bound NFeoB from S. thermophilus | | Descriptor: | 2-amino-9-(5-O-[(R)-hydroxy{[(R)-hydroxy(phosphonoamino)phosphoryl]oxy}phosphoryl]-3-O-{[2-(methylamino)phenyl]carbonyl}-beta-D-erythro-pentofuranosyl-2-ulose)-1,9-dihydro-6H-purin-6-one, Ferrous iron uptake transporter protein B, GLYCEROL, ... | | Authors: | Ash, M.R, Guilfoyle, A, Maher, M.J, Clarke, R.J, Guss, J.M, Jormakka, M. | | Deposit date: | 2010-02-24 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Potassium-activated GTPase reaction in the G Protein-coupled ferrous iron transporter B.
J.Biol.Chem., 285, 2010
|
|
3LI5
 
 | | Diisopropyl fluorophosphatase (DFPase), E21Q,N120D,N175D,D229N mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Diisopropyl-fluorophosphatase | | Authors: | Chen, J.C.-H. | | Deposit date: | 2010-01-24 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural characterization of the catalytic calcium-binding site in diisopropyl fluorophosphatase (DFPase)-Comparison with related beta-propeller enzymes.
Chem.Biol.Interact, 187, 2010
|
|
3L5B
 
 | | Structure of BACE Bound to SCH713601 | | Descriptor: | (2Z,5R)-3-(3-chlorobenzyl)-2-imino-5-methyl-5-(2-methylpropyl)imidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L6D
 
 | | Crystal structure of putative oxidoreductase from Pseudomonas putida KT2440 | | Descriptor: | Putative oxidoreductase | | Authors: | Malashkevich, V.N, Patskovsky, Y, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-12-23 | | Release date: | 2010-01-12 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of putative oxidoreductase from Pseudomonas putida KT2440
To be Published
|
|
3L6Y
 
 | | Crystal structure of p120 catenin in complex with E-cadherin | | Descriptor: | Catenin delta-1, E-cadherin | | Authors: | Ishiyama, N, Lee, S.-H, Liu, S, Li, G.-Y, Smith, M.J, Reichardt, L.F, Ikura, M. | | Deposit date: | 2009-12-27 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion.
Cell(Cambridge,Mass.), 141, 2010
|
|
3L8C
 
 | |
3L8E
 
 | | Crystal Structure of apo form of D,D-heptose 1.7-bisphosphate phosphatase from E. Coli | | Descriptor: | ACETIC ACID, D,D-heptose 1,7-bisphosphate phosphatase, ZINC ION | | Authors: | Nguyen, H, Peisach, E, Allen, K.N. | | Deposit date: | 2009-12-31 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Determinants of Substrate Recognition in the HAD Superfamily Member d-glycero-d-manno-Heptose-1,7-bisphosphate Phosphatase (GmhB) .
Biochemistry, 49, 2010
|
|
3LA5
 
 | |
3L9U
 
 | |
3LC0
 
 | |
3LCB
 
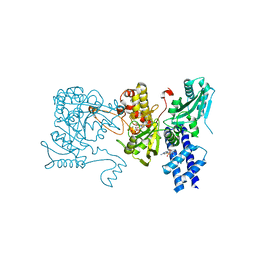 | | The crystal structure of isocitrate dehydrogenase kinase/phosphatase in complex with its substrate, isocitrate dehydrogenase, from Escherichia coli. | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Isocitrate dehydrogenase [NADP], ... | | Authors: | Zheng, J, Jia, Z. | | Deposit date: | 2010-01-10 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the bifunctional isocitrate dehydrogenase kinase/phosphatase.
Nature, 465, 2010
|
|
3LDZ
 
 | |
3LBF
 
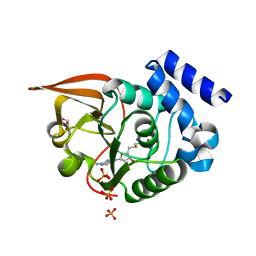 | | Crystal structure of Protein L-isoaspartyl methyltransferase from Escherichia coli | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein-L-isoaspartate O-methyltransferase, ... | | Authors: | Fang, P, Li, X, Wang, J, Niu, L, Teng, M. | | Deposit date: | 2010-01-08 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the protein L-isoaspartyl methyltransferase from Escherichia coli
Cell Biochem.Biophys., 58, 2010
|
|
3LEI
 
 | | Lectin Domain of Lectinolysin complexed with Fucose | | Descriptor: | CALCIUM ION, NICKEL (II) ION, Platelet aggregation factor Sm-hPAF, ... | | Authors: | Feil, S.C. | | Deposit date: | 2010-01-14 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the lectin regulatory domain of the cholesterol-dependent cytolysin lectinolysin reveals the basis for its lewis antigen specificity.
Structure, 20, 2012
|
|
3LET
 
 | | Crystal Structure of Fic domain containing AMPylator, VopS | | Descriptor: | Adenosine monophosphate-protein transferase vopS | | Authors: | Luong, P.H, Kinch, L.N, Brautigam, C.A, Grishin, N.V, Tomchick, D.R, Orth, K. | | Deposit date: | 2010-01-15 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural and Kinetic Analysis of VopS with Fic Domain Supports a Direct Transfer Mechanism for AMPylation
To be Published
|
|
3LE0
 
 | | Lectin Domain of Lectinolysin complexed with Glycerol | | Descriptor: | CALCIUM ION, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Feil, S.C. | | Deposit date: | 2010-01-13 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of the lectin regulatory domain of the cholesterol-dependent cytolysin lectinolysin reveals the basis for its lewis antigen specificity.
Structure, 20, 2012
|
|
3LHR
 
 | | Crystal structure of the SCAN domain from Human ZNF24 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ETHYL MERCURY ION, ... | | Authors: | Volkman, B.F, Peterson, F.C, Bingman, C.A, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-01-22 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the SCAN domain from Human ZNF24
To be Published
|
|
3LIF
 
 | |
3LGU
 
 | | Y162A mutant of the DegS-deltaPDZ protease | | Descriptor: | Protease degS | | Authors: | Sohn, J, Grant, R.A, Sauer, R.T. | | Deposit date: | 2010-01-21 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Allostery is an intrinsic property of the protease domain of DegS: implications for enzyme function and evolution.
J.Biol.Chem., 285, 2010
|
|
3LJY
 
 | |
