3DW7
 
 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23 S rRNA, U2656-SeCH3 modified | | Descriptor: | Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
3DAL
 
 | | Methyltransferase domain of human PR domain-containing protein 1 | | Descriptor: | PR domain zinc finger protein 1 | | Authors: | Amaya, M.F, Zeng, H, Antoshenko, T, Dong, A, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Plotnikov, A.N, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-29 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure
of methyltransferase domain of human PR domain-containing protein 1
To be Published
|
|
2PMR
 
 | | Crystal structure of a protein of unknown function from Methanobacterium thermoautotrophicum | | Descriptor: | Uncharacterized protein | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Wu, B, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-04-23 | | Release date: | 2007-05-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystal structure of a protein of unknown function from Methanobacterium thermoautotrophicum.
To be Published
|
|
3V3J
 
 | |
3UKQ
 
 | |
2POS
 
 | | Crystal Structure of sylvaticin, a new secreted protein from pythium sylvaticum | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Lascombe, M.B, Prange, T. | | Deposit date: | 2007-04-27 | | Release date: | 2008-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of sylvaticin, a new alpha-elicitin-like protein from Pythium sylvaticum.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3V4Z
 
 | | D-alanine--D-alanine ligase from Yersinia pestis | | Descriptor: | D-alanine--D-alanine ligase, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL | | Authors: | Osipiuk, J, Nocek, B, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-15 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | D-alanine--D-alanine ligase from Yersinia pestis.
To be Published
|
|
3V5L
 
 | |
3DHP
 
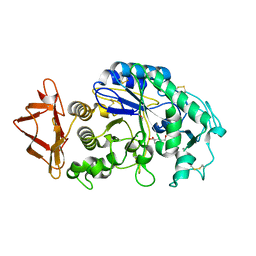 | | Probing the role of aromatic residues at the secondary saccharide binding sites of human salivary alpha-amylase in substrate hydrolysis and bacterial binding | | Descriptor: | 4-amino-4,6-dideoxy-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 5-HYDROXYMETHYL-CHONDURITOL, Alpha-amylase 1, ... | | Authors: | Ragunath, C, Manuel, S.G.A, Sait, H.M, Kasinathan, C. | | Deposit date: | 2008-06-18 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the role of aromatic residues
To be Published
|
|
2PQL
 
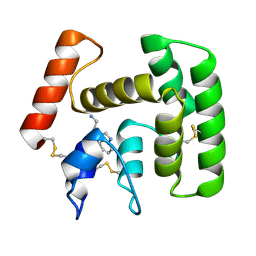 | | Crystal Structure of Anopheles gambiae D7R4-tryptamine complex | | Descriptor: | 2-(1H-INDOL-3-YL)ETHANAMINE, D7r4 protein | | Authors: | Andersen, J.F, Mans, B.J, Calvo, E, Ribeiro, J.M. | | Deposit date: | 2007-05-02 | | Release date: | 2007-10-09 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of D7r4, a Salivary Biogenic Amine-binding Protein from the Malaria Mosquito Anopheles gambiae.
J.Biol.Chem., 282, 2007
|
|
3V5T
 
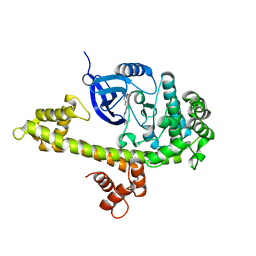 | |
3DI8
 
 | | Crystal structure of bovine pancreatic ribonuclease A variant (V57A) | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SULFATE ION | | Authors: | Kurpiewska, K, Font, J, Ribo, M, Vilanova, M, Lewinski, K. | | Deposit date: | 2008-06-20 | | Release date: | 2008-07-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic studies of RNase A variants engineered at the most destabilizing positions of the main hydrophobic core: further insight into protein stability
Proteins, 77, 2009
|
|
3V6D
 
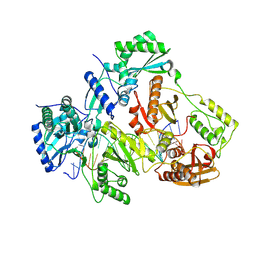 | | Crystal structure of HIV-1 reverse transcriptase (RT) cross-linked with AZT-terminated DNA | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3'), DNA (5'-D(*AP*TP*GP*GP*AP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), HIV-1 REVERSE TRANSCRIPTASE P51 subunit, ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2011-12-19 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7048 Å) | | Cite: | HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3DBZ
 
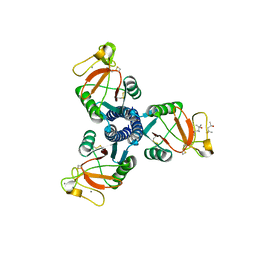 | | human surfactant protein D | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Pulmonary surfactant-associated protein D | | Authors: | Head, J.F. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interaction of recombinant surfactant protein D with lipopolysaccharide: conformation and orientation of bound protein by IRRAS and simulations.
Biochemistry, 47, 2008
|
|
3DCK
 
 | | X-ray structure of D25N chemical analogue of HIV-1 protease complexed with ketomethylene isostere inhibitor | | Descriptor: | (2S)-2-{[(2R,5S)-5-{[(2S,3S)-2-{[(2S,3R)-2-(acetylamino)-3-hydroxybutanoyl]amino}-3-methylpentanoyl]amino}-2-butyl-4-oxononanoyl]amino}-N~1~-[(2S)-1-amino-5-carbamimidamido-1-oxopentan-2-yl]pentanediamide, Chemical analogue HIV-1 protease | | Authors: | Torbeev, V.Y, Mandal, K, Terechko, V.A, Kent, S.B.H. | | Deposit date: | 2008-06-03 | | Release date: | 2008-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of chemically synthesized HIV-1 protease and a ketomethylene isostere inhibitor based on the p2/NC cleavage site
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2PFQ
 
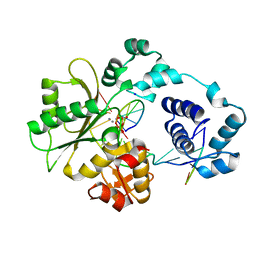 | | Manganese promotes catalysis in a DNA polymerase lambda-DNA crystal | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA polymerase lambda, Downstream Primer, ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2007-04-05 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of the catalytic metal during polymerization by DNA polymerase lambda.
DNA Repair, 6, 2007
|
|
3DCR
 
 | | X-ray structure of HIV-1 protease and hydrated form of ketomethylene isostere inhibitor | | Descriptor: | Chemical analogue HIV-1 protease, N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide | | Authors: | Torbeev, V.Y, Mandal, K, Terechko, V.A, Kent, S.B.H. | | Deposit date: | 2008-06-04 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of chemically synthesized HIV-1 protease and a ketomethylene isostere inhibitor based on the p2/NC cleavage site
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3UJN
 
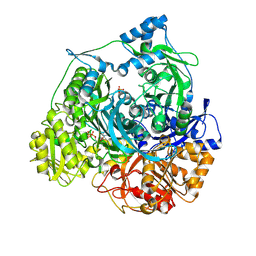 | | Formyl Glycinamide Ribonucleotide Amidotransferase from Salmonella Typhimurium : Role of the ATP complexation and glutaminase domain in catalytic coupling | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoribosylformylglycinamidine synthase, ... | | Authors: | Anand, R, Morar, M, Tanwar, A.S, Panjikar, S. | | Deposit date: | 2011-11-08 | | Release date: | 2012-06-06 | | Last modified: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Formylglycinamide ribonucleotide amidotransferase from Salmonella typhimurium: role of ATP complexation and the glutaminase domain in catalytic coupling
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3UKL
 
 | |
2PGY
 
 | | GTB C209A, no Hg | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-galactosyltransferase | | Authors: | Letts, J.A, Schuman, B. | | Deposit date: | 2007-04-10 | | Release date: | 2007-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | The effect of heavy atoms on the conformation of the active-site polypeptide loop in human ABO(H) blood-group glycosyltransferase B.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
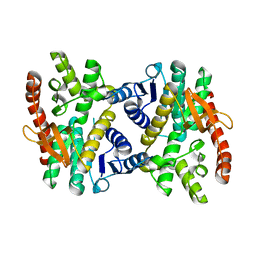
2PWZ
 
 | |
2PXD
 
 | | Variant 1 of Ribonucleoprotein Core of the E. Coli Signal Recognition Particle | | Descriptor: | 4.5 S RNA, COBALT HEXAMMINE(III), Signal recognition particle protein | | Authors: | Keel, A.Y, Rambo, R.P, Batey, R.T, Kieft, J.S. | | Deposit date: | 2007-05-14 | | Release date: | 2007-08-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A General Strategy to Solve the Phase Problem in RNA Crystallography.
Structure, 15, 2007
|
|
3UOG
 
 | | Crystal structure of putative Alcohol dehydrogenase from Sinorhizobium meliloti 1021 | | Descriptor: | Alcohol dehydrogenase, SULFATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative Alcohol dehydrogenase from Sinorhizobium meliloti 1021
To be Published
|
|
2PXP
 
 | | Variant 13 of Ribonucleoprotein Core of the E. Coli Signal Recognition Particle | | Descriptor: | 4.5 S RNA, COBALT HEXAMMINE(III), Signal recognition particle protein | | Authors: | Keel, A.Y, Rambo, R.P, Batey, R.T, Kieft, J.S. | | Deposit date: | 2007-05-14 | | Release date: | 2007-08-07 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A General Strategy to Solve the Phase Problem in RNA Crystallography.
Structure, 15, 2007
|
|
3G8R
 
 | | Crystal structure of putative spore coat polysaccharide biosynthesis protein E from Chromobacterium violaceum ATCC 12472 | | Descriptor: | Probable spore coat polysaccharide biosynthesis protein E, ZINC ION | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-12 | | Release date: | 2009-04-07 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of putative spore coat polysaccharide biosynthesis protein E from Chromobacterium violaceum ATCC 12472
To be Published
|
|
