1MLQ
 
 | |
1MLS
 
 | |
1MLG
 
 | |
1P1Q
 
 | |
4ZO1
 
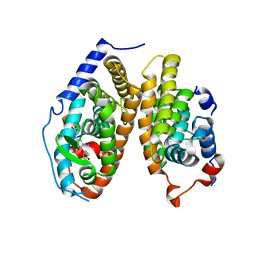 | | Crystal Structure of the T3-bound TR-beta Ligand-binding Domain in complex with RXR-alpha | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha, ... | | Authors: | Bruning, J.B, Kojetin, D.J, Matta-Camacho, E, Hughes, T.S, Srinivasan, S, Nwachukwu, J.C, Cavett, V, Nowak, J, Chalmers, M.J, Marciano, D.P, Kamenecka, T.M, Rance, M, Shulman, A.I, Mangelsdorf, D.J, Griffin, P.R, Nettles, K.W. | | Deposit date: | 2015-05-05 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.221 Å) | | Cite: | Structural mechanism for signal transduction in RXR nuclear receptor heterodimers.
Nat Commun, 6, 2015
|
|
4HR5
 
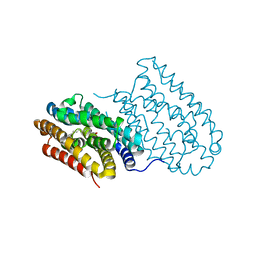 | | R2-like ligand-binding oxidase without metal cofactor | | Descriptor: | PALMITIC ACID, Ribonuleotide reductase small subunit | | Authors: | Griese, J.J, Hogbom, M. | | Deposit date: | 2012-10-26 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | Direct observation of structurally encoded metal discrimination and ether bond formation in a heterodinuclear metalloprotein
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WDK
 
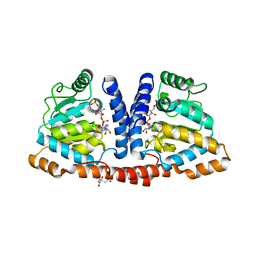 | | Crystal structure of 4-phosphopantoate-beta-alanine ligase complexed with reaction intermediate | | Descriptor: | 4-phosphopantoate--beta-alanine ligase, 5'-O-[(S)-hydroxy{[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]oxy}phosphoryl]adenosine, CITRATE ANION | | Authors: | Kishimoto, A, Kita, A, Ishibashi, T, Tomita, H, Yokooji, Y, Imanaka, T, Atomi, H, Miki, K. | | Deposit date: | 2013-06-19 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of phosphopantothenate synthetase from Thermococcus kodakarensis
Proteins, 82, 2014
|
|
1CC1
 
 | | CRYSTAL STRUCTURE OF A REDUCED, ACTIVE FORM OF THE NI-FE-SE HYDROGENASE FROM DESULFOMICROBIUM BACULATUM | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE (II) ION, HYDROGENASE (LARGE SUBUNIT), ... | | Authors: | Garcin, E, Vernede, X, Hatchikian, E.C, Volbeda, A, Frey, M, Fontecilla-Camps, J.C. | | Deposit date: | 1999-03-03 | | Release date: | 1999-06-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of a reduced [NiFeSe] hydrogenase provides an image of the activated catalytic center
Structure Fold.Des., 7, 1999
|
|
1KRC
 
 | |
3FL7
 
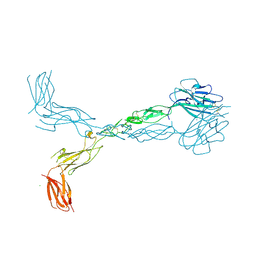 | | Crystal structure of the human ephrin A2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Ephrin receptor, ... | | Authors: | Walker, J.R, Yermekbayeva, L, Seitova, A, Butler-Cole, C, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-18 | | Release date: | 2009-01-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Architecture of Eph receptor clusters.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6H41
 
 | |
6WW3
 
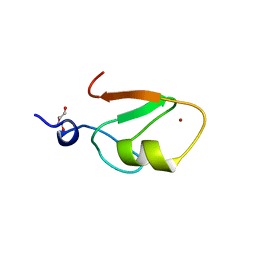 | | Crystal structure of HERC2 ZZ domain in complex with SUMO1 tail | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SUMO1 linked HERC2 ZZ domain (Small ubiquitin-related modifier 1,E3 ubiquitin-protein ligase HERC2), ... | | Authors: | Liu, J, Vann, K.R, Kutateladze, T.G. | | Deposit date: | 2020-05-07 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Structural Insight into Binding of the ZZ Domain of HERC2 to Histone H3 and SUMO1.
Structure, 28, 2020
|
|
6IOV
 
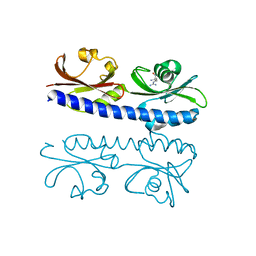 | | The ligand binding domain of Mlp37 with arginine | | Descriptor: | ARGININE, Methyl-accepting chemotaxis (MCP) signaling domain protein | | Authors: | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2018-10-31 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Structural basis of the binding affinity of chemoreceptors Mlp24p and Mlp37p for various amino acids.
Biochem.Biophys.Res.Commun., 523, 2020
|
|
1P1N
 
 | |
3QP8
 
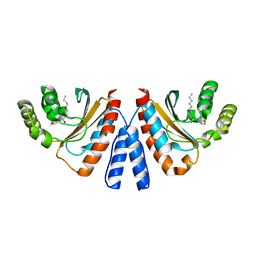 | | Crystal structure of CviR (Chromobacterium violaceum 12472) ligand-binding domain bound to C10-HSL | | Descriptor: | CviR transcriptional regulator, N-[(3S)-2-oxotetrahydrofuran-3-yl]decanamide | | Authors: | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | Deposit date: | 2011-02-11 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
7N8Y
 
 | |
4HR4
 
 | |
8FO3
 
 | |
8FO4
 
 | |
8G1F
 
 | | Structure of ACLY-D1026A-products | | Descriptor: | (3S)-citryl-Coenzyme A, ACETYL COENZYME *A, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2023-02-02 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Allosteric role of the citrate synthase homology domain of ATP citrate lyase.
Nat Commun, 14, 2023
|
|
8FO5
 
 | |
8IRC
 
 | | XFEL structure of cyanobacterial photosystem II following one flash (1F) with a 5-millisecond delay (Single conformation) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Suga, M, Shen, J.R. | | Deposit date: | 2023-03-17 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Oxygen-evolving photosystem II structures during S 1 -S 2 -S 3 transitions.
Nature, 626, 2024
|
|
8IRA
 
 | | XFEL structure of cyanobacterial photosystem II following one flash (1F) with a 200-microsecond delay | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Suga, M, Shen, J.R. | | Deposit date: | 2023-03-17 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Oxygen-evolving photosystem II structures during S 1 -S 2 -S 3 transitions.
Nature, 626, 2024
|
|
8IRI
 
 | | XFEL structure of cyanobacterial photosystem II following two flashes (2F) with a 5-millisecond delay | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Suga, M, Shen, J.R. | | Deposit date: | 2023-03-17 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Oxygen-evolving photosystem II structures during S 1 -S 2 -S 3 transitions.
Nature, 626, 2024
|
|
8G1E
 
 | | Structure of ACLY-D1026A-products-asym | | Descriptor: | (3S)-citryl-Coenzyme A, ACETYL COENZYME *A, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2023-02-02 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Allosteric role of the citrate synthase homology domain of ATP citrate lyase.
Nat Commun, 14, 2023
|
|
