1L6Y
 
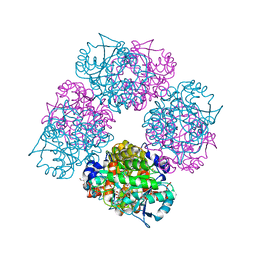 | | Crystal Structure of Porphobilinogen Synthase Complexed with the Inhibitor 4-Oxosebacic Acid | | Descriptor: | 4-OXODECANEDIOIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Jaffe, E.K, Kervinen, J, Martins, J, Stauffer, F, Neier, R, Wlodawer, A, Zdanov, A. | | Deposit date: | 2002-03-14 | | Release date: | 2002-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Species-Specific Inhibition of Porphobilinogen Synthase by 4-Oxosebacic Acid
J.Biol.Chem., 277, 2002
|
|
1L6Z
 
 | | CRYSTAL STRUCTURE OF MURINE CEACAM1A[1,4]: A CORONAVIRUS RECEPTOR AND CELL ADHESION MOLECULE IN THE CEA FAMILY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, biliary glycoprotein C | | Authors: | Tan, K, Zelus, B.D, Meijers, R, Liu, J.-H, Bergelson, J.M, Duke, N, Zhang, R, Joachimiak, A, Holmes, K.V, Wang, J.-H. | | Deposit date: | 2002-03-14 | | Release date: | 2002-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | CRYSTAL STRUCTURE OF MURINE sCEACAM1a[1,4]: A CORONAVIRUS RECEPTOR IN THE CEA FAMILY
Embo J., 21, 2002
|
|
1L70
 
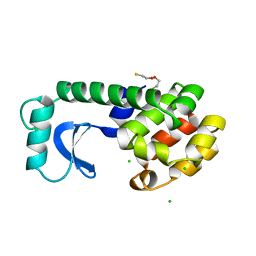 | |
1L71
 
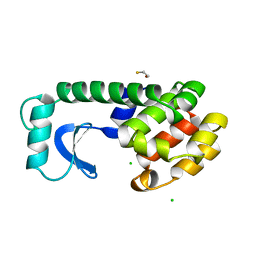 | |
1L72
 
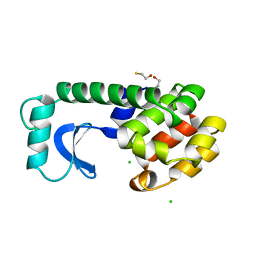 | |
1L73
 
 | |
1L74
 
 | |
1L75
 
 | |
1L76
 
 | |
1L77
 
 | |
1L79
 
 | |
1L7A
 
 | |
1L7B
 
 | | Solution NMR Structure of BRCT Domain of T. Thermophilus: Northeast Structural Genomics Consortium Target WR64TT | | Descriptor: | DNA LIGASE | | Authors: | Sahota, G, Dixon, B.L, Huang, Y.P, Aramini, J, Monleon, D, Bhattacharya, D, Swapna, G.V.T, Yin, C, Xiao, R, Anderson, S, Tejero, R, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-03-14 | | Release date: | 2003-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Brct Domain from Thermus Thermophilus DNA Ligase
To be Published
|
|
1L7C
 
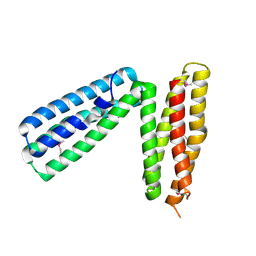 | | alpha-catenin fragment, residues 385-651 | | Descriptor: | Alpha E-catenin | | Authors: | Pokutta, S, Drees, F, Takai, Y, Nelson, W.J, Weis, W.I. | | Deposit date: | 2002-03-14 | | Release date: | 2002-06-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical and structural definition of the l-afadin- and actin-binding sites of alpha-catenin.
J.Biol.Chem., 277, 2002
|
|
1L7D
 
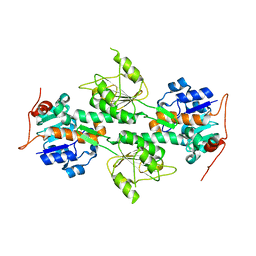 | | Crystal Structure of R. rubrum Transhydrogenase Domain I without Bound NAD(H) | | Descriptor: | nicotinamide nucleotide Transhydrogenase, subunit alpha 1 | | Authors: | Prasad, G.S, Wahlberg, M, Sridhar, V, Yamaguchi, M, Hatefi, Y, Stout, C.D. | | Deposit date: | 2002-03-14 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structures of Transhydrogenase Domain I
with and without Bound NADH
Biochemistry, 41, 2002
|
|
1L7E
 
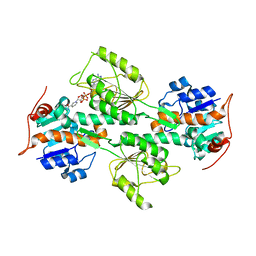 | | Crystal Structure of R. rubrum Transhydrogenase Domain I with Bound NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, nicotinamide nucleotide Transhydrogenase, subunit alpha 1 | | Authors: | Prasad, G.S, Wahlberg, M, Sridhar, V, Yamaguchi, M, Hatefi, Y, Stout, C.D. | | Deposit date: | 2002-03-14 | | Release date: | 2002-11-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Transhydrogenase Domain I
with and without Bound NADH
Biochemistry, 41, 2002
|
|
1L7F
 
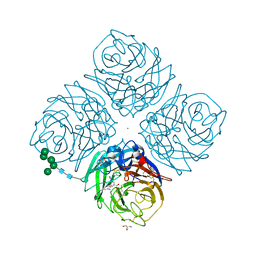 | | Crystal structure of influenza virus neuraminidase in complex with BCX-1812 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, ... | | Authors: | Smith, B.J, McKimm-Breshkin, J.L, McDonald, M, Fernley, R.T, Varghese, J.N, Colman, P.M. | | Deposit date: | 2002-03-15 | | Release date: | 2002-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of the resistance of influenza virus neuramindase to inhibitors.
J.Med.Chem., 45, 2002
|
|
1L7G
 
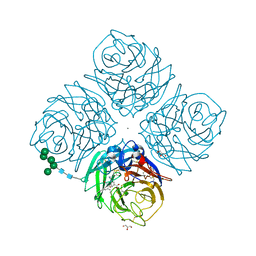 | | Crystal structure of E119G mutant influenza virus neuraminidase in complex with BCX-1812 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, ... | | Authors: | Smith, B.J, McKimm-Breshkin, J.L, McDonald, M, Fernley, R.T, Varghese, J.N, Colman, P.M. | | Deposit date: | 2002-03-15 | | Release date: | 2002-05-29 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural studies of the resistance of influenza virus neuramindase to inhibitors.
J.Med.Chem., 45, 2002
|
|
1L7H
 
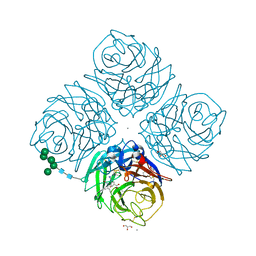 | | Crystal structure of R292K mutant influenza virus neuraminidase in complex with BCX-1812 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, ... | | Authors: | Smith, B.J, McKimm-Breshkin, J.L, McDonald, M, Fernley, R.T, Varghese, J.N, Colman, P.M. | | Deposit date: | 2002-03-15 | | Release date: | 2002-05-29 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural studies of the resistance of influenza virus neuramindase to inhibitors.
J.Med.Chem., 45, 2002
|
|
1L7I
 
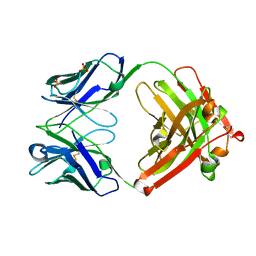 | | Crystal Structure of the anti-ErbB2 Fab2C4 | | Descriptor: | SULFATE ION, chimera of Fab2C4: "humanized" murine monoclonal antibody | | Authors: | Vajdos, F.F, Adams, C.W, Breece, T.N, Presta, L.G, de Vos, A.M, Sidhu, S.S. | | Deposit date: | 2002-03-15 | | Release date: | 2002-07-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comprehensive functional maps of the antigen-binding site of an anti-ErbB2 antibody obtained with shotgun scanning mutagenesis.
J.Mol.Biol., 320, 2002
|
|
1L7J
 
 | |
1L7K
 
 | |
1L7L
 
 | | Crystal structure of Pseudomonas aeruginosa lectin 1 determined by single wavelength anomalous scattering phasing method | | Descriptor: | CALCIUM ION, PA-I galactophilic lectin | | Authors: | Liu, Z.J, Tempel, W, Lin, D, Karaveg, K, Doyle, R.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2002-03-15 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure determination of P. aeruginosa lectin-1 using single
wavelength anomalous scattering data from native crystals (P028)
AM.CRYST.ASSOC.,ABSTR.PAPERS (ANNUAL MEETING), 29, 2002
|
|
1L7M
 
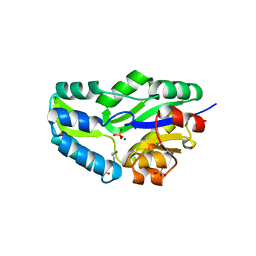 | | HIGH RESOLUTION LIGANDED STRUCTURE OF PHOSPHOSERINE PHOSPHATASE (PI COMPLEX) | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Phosphoserine Phosphatase | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-15 | | Release date: | 2002-04-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
1L7N
 
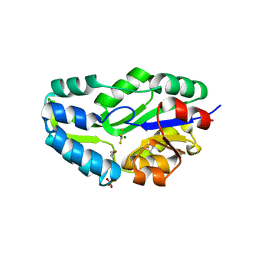 | | TRANSITION STATE ANALOGUE OF PHOSPHOSERINE PHOSPHATASE (ALUMINUM FLUORIDE COMPLEX) | | Descriptor: | ALUMINUM FLUORIDE, MAGNESIUM ION, PHOSPHOSERINE PHOSPHATASE, ... | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-16 | | Release date: | 2002-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
