5QOY
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with YW-FY-378 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(2H-1,3-benzodioxole-5-carbonyl)-2,3,4,5-tetrahydro-1H-1,4-diazepin-1-yl]ethan-1-one, ACETATE ION, ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7G16
 
 | | Crystal Structure of human FABP4 in complex with 6-bromo-3-hexyl-2-oxo-3,4-dihydro-1H-quinoline-4-carboxylic acid | | Descriptor: | (3S,4R)-6-bromo-3-hexyl-2-oxo-1,2,3,4-tetrahydroquinoline-4-carboxylic acid, BROMIDE ION, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Hofheinz, W, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
7FXY
 
 | | Crystal Structure of human FABP4 in complex with (4-chlorophenyl)methyl 5-fluoro-2,4-dioxo-1H-pyrimidine-6-carboxylate | | Descriptor: | (4-chlorophenyl)methyl 5-fluoro-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylate, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Kiss, J, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
2O4S
 
 | | Crystal Structure of HIV-1 Protease (Q7K) in Complex with Lopinavir | | Descriptor: | CHLORIDE ION, GLYCEROL, N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, ... | | Authors: | Armstrong, A.A, Muzammil, S, Jakalian, A, Bonneau, P.R, Schmelmer, V, Freire, E, Amzel, L.M. | | Deposit date: | 2006-12-04 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Unique thermodynamic response of tipranavir to human immunodeficiency virus type 1 protease drug resistance mutations.
J.Virol., 81, 2007
|
|
1RBY
 
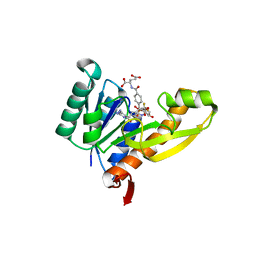 | | Human GAR Tfase complex structure with 10-(trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic acid and substrate beta-GAR | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-{4-[(1R)-4-[(2R,4R,5S)-2,4-DIAMINO-6-OXOHEXAHYDROPYRIMIDIN-5-YL]-1-(2,2,2-TRIFLUORO-1,1-DIHYDROXYETHYL)BUTYL]BENZOYL}-D-GLUTAMIC ACID, PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Zhang, Y, Desharnais, J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2003-11-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Human GAR Tfase complex structure
To be Published
|
|
2OGN
 
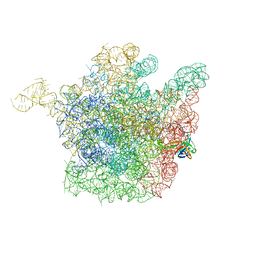 | | The crystal structure of the large ribosomal subunit from Deinococcus radiodurans complexed with the pleuromutilin derivative SB-280080 | | Descriptor: | (3AS,4R,5S,6S,8R,9R,9AR,10R)-5-HYDROXY-4,6,9,10-TETRAMETHYL-1-OXO-6-VINYLDECAHYDRO-3A,9-PROPANOCYCLOPENTA[8]ANNULEN-8-YL (PIPERIDIN-4-YLTHIO)ACETATE, 23S ribosomal RNA, 50S ribosomal protein L3 | | Authors: | Davidovich, C, Bashan, A, Auerbach-Nevo, T, Yonath, A. | | Deposit date: | 2007-01-07 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Induced-fit tightens pleuromutilins binding to ribosomes and remote interactions enable their selectivity.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7FZM
 
 | | Crystal Structure of human FABP4 in complex with N-(2,4-dichlorophenyl)-2-(2,6-dihydroxypyrimidin-4-yl)acetamide | | Descriptor: | Fatty acid-binding protein, adipocyte, N-(2,4-dichlorophenyl)-2-(2,6-dioxo-1,2,5,6-tetrahydropyrimidin-4-yl)acetamide, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Obst-Sander, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
2NTH
 
 | | Structure of Spin-labeled T4 Lysozyme Mutant L118R1 | | Descriptor: | Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2006-11-07 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme.
Protein Sci., 16, 2007
|
|
7FXA
 
 | | Crystal Structure of human FABP4 in complex with 6-bromo-4-hexyl-2-oxo-1,3-dihydroquinoline-4-carboxylic acid | | Descriptor: | (4S)-6-bromo-4-hexyl-2-oxo-1,2,3,4-tetrahydroquinoline-4-carboxylic acid, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Hofheinz, W, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
3B86
 
 | |
5ODH
 
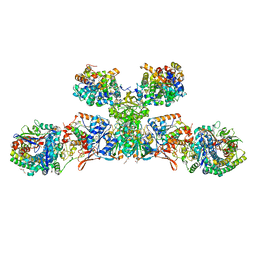 | | Heterodisulfide reductase / [NiFe]-hydrogenase complex from Methanothermococcus thermolithotrophicus soaked with heterodisulfide for 3.5 minutes | | Descriptor: | 1-THIOETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | Authors: | Wagner, T, Koch, J, Ermler, U, Shima, S. | | Deposit date: | 2017-07-05 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction.
Science, 357, 2017
|
|
7GZS
 
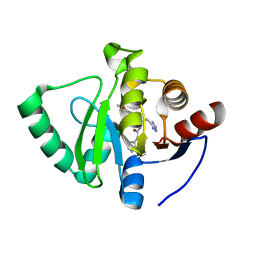 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0008340-001 | | Descriptor: | N-[(1R)-1-(4-methoxyphenyl)-2-(1H-tetrazol-5-yl)ethyl]-7H-pyrrolo[2,3-d]pyrimidine-4-carboxamide, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
3MXO
 
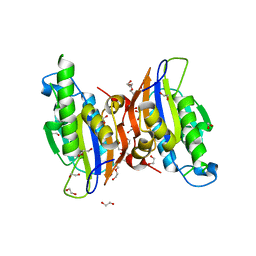 | | Crystal structure oh human phosphoglycerate mutase family member 5 (PGAM5) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chaikuad, A, Alfano, I, Picaud, S, Filippakopoulos, P, Barr, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Takeda, K, Ichijo, H, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-07 | | Release date: | 2010-09-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of PGAM5 Provide Insight into Active Site Plasticity and Multimeric Assembly.
Structure, 25, 2017
|
|
7H07
 
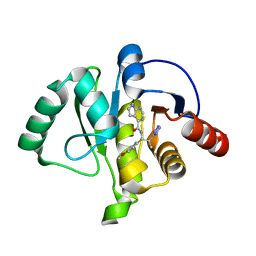 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0010744-001 | | Descriptor: | N-[(1S,2S)-1-(4-methoxyphenyl)-2-(1H-tetrazol-5-yl)propyl]-7H-pyrrolo[2,3-d]pyrimidine-4-carboxamide, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
3BH7
 
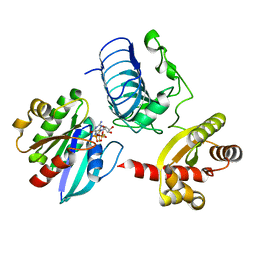 | | Crystal structure of the RP2-Arl3 complex bound to GDP-AlF4 | | Descriptor: | ADP-ribosylation factor-like protein 3, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Veltel, S, Gasper, R, Wittinghofer, A. | | Deposit date: | 2007-11-28 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The retinitis pigmentosa 2 gene product is a GTPase-activating protein for Arf-like 3
Nat.Struct.Mol.Biol., 15, 2008
|
|
7H1F
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0015381-001 | | Descriptor: | N-[(1R)-1-(4-methoxyphenyl)-2-(1H-tetrazol-5-yl)ethyl]-7H-pyrrolo[2,3-d]pyrimidine-4-carboxamide, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
3MYG
 
 | | Aurora A Kinase complexed with SCH 1473759 | | Descriptor: | 2-{ethyl[(5-{[6-methyl-3-(1H-pyrazol-4-yl)imidazo[1,2-a]pyrazin-8-yl]amino}isothiazol-3-yl)methyl]amino}-2-methylpropan-1-ol, Serine/threonine-protein kinase 6, TETRAETHYLENE GLYCOL | | Authors: | Hruza, A, Prosis, W, Ramanathan, L. | | Deposit date: | 2010-05-10 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Potent, Injectable Inhibitor of Aurora Kinases Based on the Imidazo-[1,2-a]-Pyrazine Core.
ACS Med Chem Lett, 1, 2010
|
|
5O3N
 
 | | Crystal structure of E. cloacae 3,4-dihydroxybenzoic acid decarboxylase (AroY) reconstituted with prFMN | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, 3,4-dihydroxybenzoate decarboxylase, GLYCEROL, ... | | Authors: | Marshall, S.A, Leys, D. | | Deposit date: | 2017-05-24 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Regioselective para-Carboxylation of Catechols with a Prenylated Flavin Dependent Decarboxylase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
1TFT
 
 | | NMR Structure of an Antagonists of the XIAP-Caspase-9 Interaction Complexed to the BIR3 domain of XIAP | | Descriptor: | 1-[3,3-DIMETHYL-2-(2-METHYLAMINO-PROPIONYLAMINO)-BUTYRYL]-4-PHENOXY-PYRROLIDINE-2-CARBOXYLIC ACID(1,2,3,4-TETRAHYDRO-NAPHTHALEN-1-YL)-AMIDE, Baculoviral IAP repeat-containing protein 4, ZINC ION | | Authors: | Oost, T.K, Sun, C, Armstrong, R.C, Al-Assaad, A.S, Betz, S.F, Deckwerth, T.L, Elmore, S.W, Meadows, R.P, Olejniczak, E.T, Oleksijew, A.K, Oltersdorf, T, Rosenberg, S.H, Shoemaker, A.R, Zou, H, Fesik, S.W. | | Deposit date: | 2004-05-27 | | Release date: | 2005-05-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Discovery of potent antagonists of the antiapoptotic protein XIAP for the treatment of cancer.
J.Med.Chem., 47, 2004
|
|
7GE7
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ADA-UCB-6c2cb422-3 (Mpro-x11354) | | Descriptor: | 2-(4-methylpyridin-3-yl)-N-(1,2,3,4-tetrahydroisoquinolin-8-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
3ATQ
 
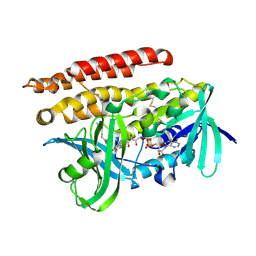 | | Geranylgeranyl Reductase (GGR) from Sulfolobus acidocaldarius | | Descriptor: | Conserved Archaeal protein, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, TETRADECANE | | Authors: | Sasaki, D, Fujihashi, M, Murakami, M, Yoshimura, T, Hemmi, H, Miki, K. | | Deposit date: | 2011-01-12 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and mutation analysis of archaeal geranylgeranyl reductase
J.Mol.Biol., 409, 2011
|
|
7GGR
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-c7771779-1 (Mpro-x12695) | | Descriptor: | (4S)-6-chloro-4-hydroxy-N-(isoquinolin-4-yl)-2-oxo-1,2,3,4-tetrahydroquinoline-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
3SAO
 
 | | The Siderocalin Ex-FABP functions through dual ligand specificities | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, 2,3-DIHYDROXY-BENZOIC ACID, Extracellular fatty acid-binding protein, ... | | Authors: | Correnti, C, Strong, R.K, Clifton, M.C. | | Deposit date: | 2011-06-03 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Galline Ex-FABP Is an Antibacterial Siderocalin and a Lysophosphatidic Acid Sensor Functioning through Dual Ligand Specificities.
Structure, 19, 2011
|
|
7GGQ
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-92e193ae-1 (Mpro-x12692) | | Descriptor: | (4R)-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroquinoline-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.811 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GGV
 
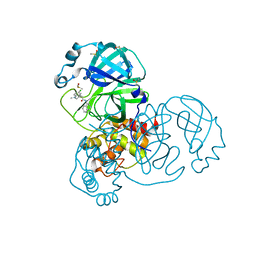 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MIC-UNK-91acba05-6 (Mpro-x12710) | | Descriptor: | (4R)-6-chloro-N-(isoquinolin-4-yl)-1-methyl-1,2,3,4-tetrahydroquinoline-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
