1AXI
 
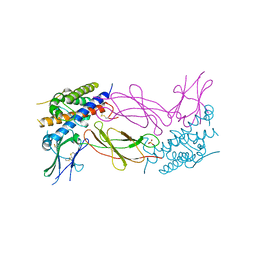 | | STRUCTURAL PLASTICITY AT THE HGH:HGHBP INTERFACE | | Descriptor: | GROWTH HORMONE, GROWTH HORMONE RECEPTOR, SULFATE ION | | Authors: | Atwell, S, Ultsch, M, De Vos, A.M, Wells, J.A. | | Deposit date: | 1997-10-15 | | Release date: | 1998-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural plasticity in a remodeled protein-protein interface.
Science, 278, 1997
|
|
1AXJ
 
 | |
1AXK
 
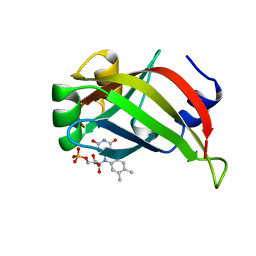
 | | ENGINEERED BACILLUS BIFUNCTIONAL ENZYME GLUXYN-1 | | Descriptor: | CALCIUM ION, GLUXYN-1 | | Authors: | Ay, J, Heinemann, U. | | Deposit date: | 1997-10-16 | | Release date: | 1999-05-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of the Bacillus hybrid enzyme GluXyn-1: native-like jellyroll fold preserved after insertion of autonomous globular domain.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1AXL
 
 | | SOLUTION CONFORMATION OF THE (-)-TRANS-ANTI-[BP]DG ADDUCT OPPOSITE A DELETION SITE IN DNA DUPLEX D(CCATC-[BP]G-CTACC)D(GGTAG--GATGG), NMR, 6 STRUCTURES | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(CCATC-[BP]G-CTACC)D(GGTAG--GATGG) | | Authors: | Feng, B, Gorin, A.A, Kolbanovskiy, A, Hingerty, B.E, Geacintov, N.E, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-16 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of the (-)-trans-anti-[BP]dG adduct opposite a deletion site in a DNA duplex: intercalation of the covalently attached benzo[a]pyrene into the helix with base displacement of the modified deoxyguanosine into the minor groove.
Biochemistry, 36, 1997
|
|
1AXM
 
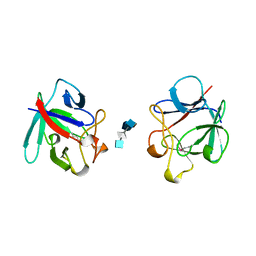 | | HEPARIN-LINKED BIOLOGICALLY-ACTIVE DIMER OF FIBROBLAST GROWTH FACTOR | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ACIDIC FIBROBLAST GROWTH FACTOR | | Authors: | DiGabriele, A.D, Lax, I, Chen, D.I, Svahn, C.M, Jaye, M, Schlessinger, J, Hendrickson, W.A. | | Deposit date: | 1997-10-16 | | Release date: | 1998-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a heparin-linked biologically active dimer of fibroblast growth factor.
Nature, 393, 1998
|
|
1AXN
 
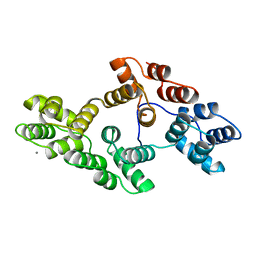 | |
1AXO
 
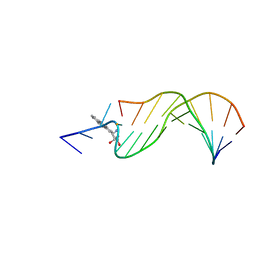 | | STRUCTURAL ALIGNMENT OF THE (+)-TRANS-ANTI-[BP]DG ADDUCT POSITIONED OPPOSITE DC AT A DNA TEMPLATE-PRIMER JUNCTION, NMR, 6 STRUCTURES | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(AAC-[BP]G-CTACCATCC)D(GGATGGTAGC) | | Authors: | Feng, B, Gorin, A.A, Hingerty, B.E, Geacintov, N.E, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-16 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural alignment of the (+)-trans-anti-benzo[a]pyrene-dG adduct positioned opposite dC at a DNA template-primer junction.
Biochemistry, 36, 1997
|
|
1AXP
 
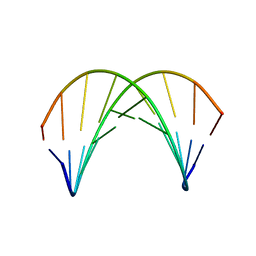 | | DNA DUPLEX CONTAINING A PURINE-RICH STRAND, NMR, 6 STRUCTURES | | Descriptor: | DNA (D(GAAGAGAAGC)(DOT)D(GCTTCTCTTC)) | | Authors: | Gyi, J.I, Lane, A.N, Conn, G.L, Brown, T. | | Deposit date: | 1997-10-17 | | Release date: | 1998-04-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of DNA.RNA hybrids with purine-rich and pyrimidine-rich strands: comparison with the homologous DNA and RNA duplexes.
Biochemistry, 37, 1998
|
|
1AXQ
 
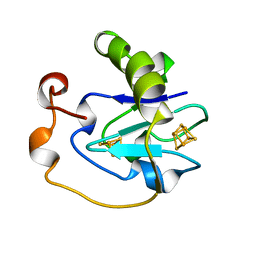 | | FERRICYANIDE OXIDIZED FDI | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Stout, C.D. | | Deposit date: | 1997-10-17 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of ferricyanide-oxidized [Fe-S] clusters in Azotobacter vinelandii ferredoxin I.
J.Biol.Inorg.Chem., 3, 1998
|
|
1AXR
 
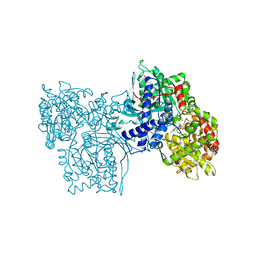 | | COOPERATIVITY BETWEEN HYDROGEN-BONDING AND CHARGE-DIPOLE INTERACTIONS IN THE INHIBITION OF BETA-GLYCOSIDASES BY AZOLOPYRIDINES: EVIDENCE FROM A STUDY WITH GLYCOGEN PHOSPHORYLASE B | | Descriptor: | 4,5,6-TRIHYDROXY-7-HYDROXYMETHYL-4,5,6,7-TETRAHYDRO-1H-[1,2,3]TRIAZOLO[1,5-A]PYRIDIN-8-YLIUM, GLYCOGEN PHOSPHORYLASE, PHOSPHATE ION, ... | | Authors: | Oikonomakos, N.G, Heightman, T.D. | | Deposit date: | 1997-10-20 | | Release date: | 1998-01-28 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cooperative interactions of the catalytic nucleophile and the catalytic acid in the inhibition of beta-glycosidases. Calculations and their validation by comparative kinetic and structural studies of the inhibition of glycogen phosphorylase b.
HELV.CHIM.ACTA, 81, 1998
|
|
1AXS
 
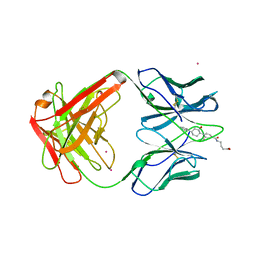 | | MATURE OXY-COPE CATALYTIC ANTIBODY WITH HAPTEN | | Descriptor: | (1S,2S,5S)2-(4-GLUTARIDYLBENZYL)-5-PHENYL-1-CYCLOHEXANOL, CADMIUM ION, OXY-COPE CATALYTIC ANTIBODY | | Authors: | Mundorff, E.C, Ulrich, H.D, Stevens, R.C. | | Deposit date: | 1997-10-20 | | Release date: | 1998-02-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The interplay between binding energy and catalysis in the evolution of a catalytic antibody.
Nature, 389, 1997
|
|
1AXT
 
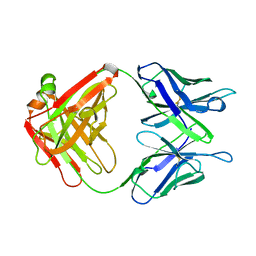 | |
1AXU
 
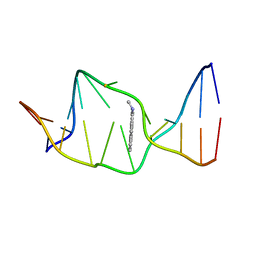 | | SOLUTION NMR STRUCTURE OF THE [AP]DG ADDUCT OPPOSITE DA IN A DNA DUPLEX, NMR, 9 STRUCTURES | | Descriptor: | DNA DUPLEX D(CCATC-[AP]G-CTACC)D(GGTAGAGATGG), N-1-AMINOPYRENE | | Authors: | Gu, Z, Gorin, A.A, Krishnasami, R, Hingerty, B.E, Basu, A.K, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-21 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-(deoxyguanosin-8-yl)-1-aminopyrene ([AP]dG) adduct opposite dA in a DNA duplex.
Biochemistry, 38, 1999
|
|
1AXV
 
 | | SOLUTION NMR STRUCTURE OF THE [BP]DA ADDUCT OPPOSITE DT IN A DNA DUPLEX, 6 STRUCTURES | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(CTCTC-[BP]A-CTTCC)D(GGAAGTGAGAG) | | Authors: | Mao, B, Gu, Z, Gorin, A.A, Chen, J, Hingerty, B.E, Amid, S, Broyde, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 1997-10-21 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the (+)-cis-anti-benzo[a]pyrene-dA ([BP]dA) adduct opposite dT in a DNA duplex.
Biochemistry, 38, 1999
|
|
1AXW
 
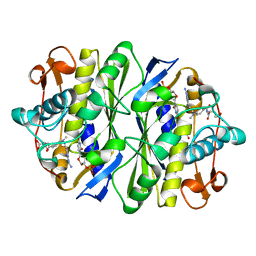 | |
1AXX
 
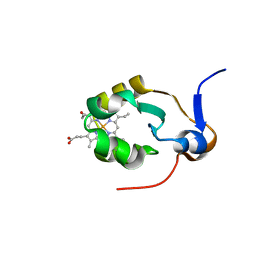 | | THE SOLUTION STRUCTURE OF OXIDIZED RAT MICROSOMAL CYTOCHROME B5, NMR, 19 STRUCTURES | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Arnesano, F, Banci, L, Bertini, I, Felli, I.C. | | Deposit date: | 1997-10-22 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of oxidized rat microsomal cytochrome b5.
Biochemistry, 37, 1998
|
|
1AXY
 
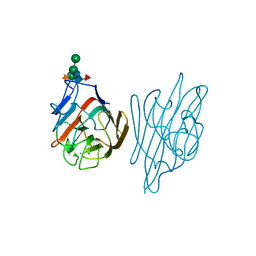 | | ERYTHRINA CORALLODENDRON LECTIN | | Descriptor: | CALCIUM ION, LECTIN, MANGANESE (II) ION, ... | | Authors: | Shaanan, B, Elgavish, S. | | Deposit date: | 1997-10-24 | | Release date: | 1998-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Erythrina corallodendron lectin and of its complexes with mono- and disaccharides.
J.Mol.Biol., 277, 1998
|
|
1AXZ
 
 | |
1AY0
 
 | | IDENTIFICATION OF CATALYTICALLY IMPORTANT RESIDUES IN YEAST TRANSKETOLASE | | Descriptor: | CALCIUM ION, THIAMINE DIPHOSPHATE, TRANSKETOLASE | | Authors: | Wikner, C, Nilsson, U, Meshalkina, L, Udekwu, C, Lindqvist, Y, Schneider, G. | | Deposit date: | 1997-11-13 | | Release date: | 1998-05-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of catalytically important residues in yeast transketolase.
Biochemistry, 36, 1997
|
|
1AY1
 
 | | ANTI TAQ FAB TP7 | | Descriptor: | TP7 FAB | | Authors: | Murali, R, Helmer-Citterich, M, Sharkey, D.J, Scalice, E.R, Daiss, J.L, Sullivan, M.A, Murthy, H.M.K. | | Deposit date: | 1997-11-13 | | Release date: | 1998-05-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural studies on an inhibitory antibody against Thermus aquaticus DNA polymerase suggest mode of inhibition.
Protein Eng., 11, 1998
|
|
1AY2
 
 | | STRUCTURE OF THE FIBER-FORMING PROTEIN PILIN AT 2.6 ANGSTROMS RESOLUTION | | Descriptor: | HEPTANE-1,2,3-TRIOL, PLATINUM (II) ION, TYPE 4 PILIN, ... | | Authors: | Forest, K.T, Parge, H.E, Tainer, J.A. | | Deposit date: | 1997-11-13 | | Release date: | 1998-04-29 | | Last modified: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the fibre-forming protein pilin at 2.6 A resolution.
Nature, 378, 1995
|
|
1AY3
 
 | | Nodularin from Nodularia spumigena | | Descriptor: | PEPTIDIC TOXIN NODULARIN | | Authors: | Annila, A.J. | | Deposit date: | 1997-11-14 | | Release date: | 1999-08-16 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of nodularin. An inhibitor of serine/threonine-specific protein phosphatases.
J.Biol.Chem., 271, 1996
|
|
1AY4
 
 | | AROMATIC AMINO ACID AMINOTRANSFERASE WITHOUT SUBSTRATE | | Descriptor: | AROMATIC AMINO ACID AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Okamoto, A, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 1997-11-14 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structures of Paracoccus denitrificans aromatic amino acid aminotransferase: a substrate recognition site constructed by rearrangement of hydrogen bond network.
J.Mol.Biol., 280, 1998
|
|
1AY5
 
 | | AROMATIC AMINO ACID AMINOTRANSFERASE COMPLEX WITH MALEATE | | Descriptor: | AROMATIC AMINO ACID AMINOTRANSFERASE, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Okamoto, A, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 1997-11-14 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Paracoccus denitrificans aromatic amino acid aminotransferase: a substrate recognition site constructed by rearrangement of hydrogen bond network.
J.Mol.Biol., 280, 1998
|
|
1AY6
 
 | | THROMBIN INHIBITOR FROM THEONALLA, CYCLOTHEANAMIDE-BASED MACROCYCLIC TRIPEPTIDE MOTIF | | Descriptor: | HIRUGEN, THROMBIN HEAVY CHAIN, THROMBIN LIGHT CHAIN, ... | | Authors: | Ganesh, V, Maryanoff, B.E, Tulinsky, A. | | Deposit date: | 1997-11-14 | | Release date: | 1998-03-18 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel thrombin inhibitors that are based on a macrocyclic tripeptide motif
Bioorg.Med.Chem.Lett., 6, 1996
|
|
