5XTZ
 
 | | Crystal structure of GAS41 YEATS bound to H3K27ac peptide | | Descriptor: | ACETATE ION, THR-LYS-ALA-ALA-ARG-ALY-SER-ALA-PRO-ALA, YEATS domain-containing protein 4 | | Authors: | Li, H.T, Zhao, D. | | Deposit date: | 2017-06-21 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Gas41 links histone acetylation to H2A.Z deposition and maintenance of embryonic stem cell identity.
Cell Discov, 4, 2018
|
|
2GOF
 
 | | Three-dimensional structure of the trans-membrane domain of Vpu from HIV-1 in aligned phospholipid bicelles | | Descriptor: | VPU protein | | Authors: | Park, S.H, De Angelis, A.A, Nevzorov, A.A, Wu, C.H, Opella, S.J. | | Deposit date: | 2006-04-12 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Structure of the Transmembrane Domain of Vpu from HIV-1 in Aligned Phospholipid Bicelles.
Biophys.J., 91, 2006
|
|
5AJZ
 
 | | Human PFKFB3 in complex with an indole inhibitor 5 | | Descriptor: | 2-azanyl-N-[4-[(3-cyano-1H-indol-5-yl)oxy]phenyl]ethanamide, 6-O-phosphono-beta-D-fructofuranose, 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE 3, ... | | Authors: | Boyd, S, Brookfield, J.L, Critchlow, S.E, Cumming, I.A, Curtis, N.J, Debreczeni, J.E, Degorce, S.L, Donald, C, Evans, N.J, Groombridge, S, Hopcroft, P, Jones, N.P, Kettle, J.G, Lamont, S, Lewis, H.J, MacFaull, P, McLoughlin, S.B, Rigoreau, L.J.M, Smith, J.M, St-Gallay, S, Stock, J.K, Wheatley, E.R, Winter, J, Wingfield, J. | | Deposit date: | 2015-02-27 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-Based Design of Potent and Selective Inhibitors of the Metabolic Kinase Pfkfb3.
J.Med.Chem., 58, 2015
|
|
5AK0
 
 | | Human PFKFB3 in complex with an indole inhibitor 6 | | Descriptor: | (2S)-N-[4-[1-METHYL-3-(1-METHYLPYRAZOL-4-YL)INDOL-5-YL]OXYPHENYL]PYRROLIDINE-2-CARBOXAMIDE, 6-O-phosphono-beta-D-fructofuranose, 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE 3, ... | | Authors: | Boyd, S, Brookfield, J.L, Critchlow, S.E, Cumming, I.A, Curtis, N.J, Debreczeni, J.E, Degorce, S.L, Donald, C, Evans, N.J, Groombridge, S, Hopcroft, P, Jones, N.P, Kettle, J.G, Lamont, S, Lewis, H.J, MacFaull, P, McLoughlin, S.B, Rigoreau, L.J.M, Smith, J.M, St-Gallay, S, Stock, J.K, Wheatley, E.R, Winter, J, Wingfield, J. | | Deposit date: | 2015-02-27 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure-Based Design of Potent and Selective Inhibitors of the Metabolic Kinase Pfkfb3.
J.Med.Chem., 58, 2015
|
|
4HFZ
 
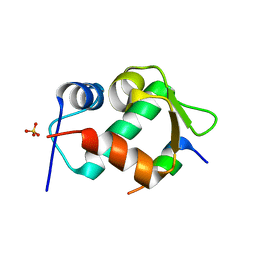 | | Crystal Structure of an MDM2/P53 Peptide Complex | | Descriptor: | Cellular tumor antigen p53, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Anil, B, Riedinger, C, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | The structure of an MDM2-Nutlin-3a complex solved by the use of a validated MDM2 surface-entropy reduction mutant.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3GU5
 
 | |
2I3W
 
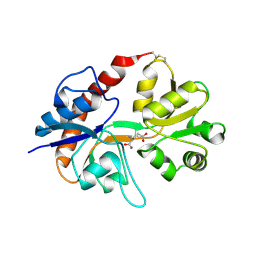 | | Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor: Structure of S729C mutant | | Descriptor: | GLUTAMATE RECEPTOR SUBUNIT 2, GLUTAMIC ACID | | Authors: | Armstrong, N, Jasti, J, Beich-Frandsen, M, Gouaux, E. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Measurement of Conformational Changes accompanying Desensitization in an Ionotropic Glutamate Receptor.
Cell(Cambridge,Mass.), 127, 2006
|
|
1GQY
 
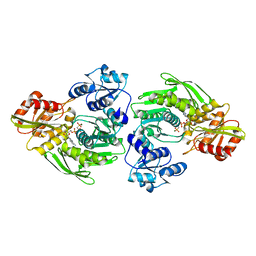 | | MURC - CRYSTAL STRUCTURE OF THE ENZYME FROM HAEMOPHILUS INFLUENZAE COMPLEXED WITH AMPPCP | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, UDP-N-ACETYLMURAMATE-L-ALANINE LIGASE | | Authors: | Skarzynski, T, Cleasby, A, Domenici, E, Gevi, M, Shaw, J. | | Deposit date: | 2001-12-06 | | Release date: | 2003-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Udp-N-Acetylmuramate-L-Alanine Ligase (Murc) from Haemophilus Influenzae
To be Published
|
|
1DRA
 
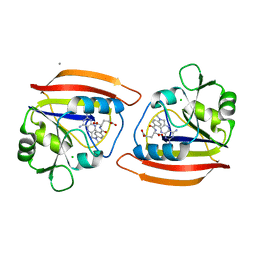 | |
1DR6
 
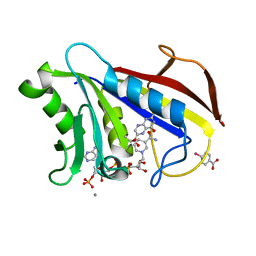 | | CRYSTAL STRUCTURES OF ORGANOMERCURIAL-ACTIVATED CHICKEN LIVER DIHYDROFOLATE REDUCTASE COMPLEXES | | Descriptor: | 7,8-DIHYDROBIOPTERIN, CALCIUM ION, DIHYDROFOLATE REDUCTASE, ... | | Authors: | Mctigue, M.A, Davies /II, J.F, Kaufman, B.T, Xuong, N.-H, Kraut, J. | | Deposit date: | 1992-03-14 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Organomercurial-Activated Chicken Liver Dihydrofolate Reductase Complexes
To be Published
|
|
1GR2
 
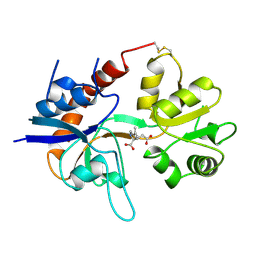 | | STRUCTURE OF A GLUTAMATE RECEPTOR LIGAND BINDING CORE (GLUR2) COMPLEXED WITH KAINATE | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, PROTEIN (GLUTAMATE RECEPTOR 2) | | Authors: | Armstrong, N, Sun, Y, Chen, G.Q, Gouaux, E. | | Deposit date: | 1998-09-17 | | Release date: | 1998-12-09 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a glutamate-receptor ligand-binding core in complex with kainate.
Nature, 395, 1998
|
|
1GZ2
 
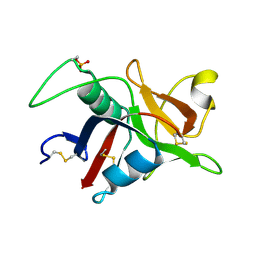 | |
2JOQ
 
 | | Solution Structure of Protein HP0495 from H. pylori; Northeast structural genomics consortium target PT2; Ontario Centre for Structural Proteomics target HP0488 | | Descriptor: | Hypothetical protein HP_0495 | | Authors: | Gutmanas, A, Lemak, A, Yee, A, Karra, M, Srisailam, S, Semesi, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2007-03-19 | | Release date: | 2007-04-03 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Protein HP0495 from H. pylori
To be Published
|
|
2E21
 
 | | Crystal structure of TilS in a complex with AMPPNP from Aquifex aeolicus. | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, tRNA(Ile)-lysidine synthase | | Authors: | Kuratani, M, Yoshikawa, Y, Sekine, S, Ishii, T, Shibata, R, Bessho, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-11-06 | | Release date: | 2007-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the initial binding of tRNA(Ile) lysidine synthetase TilS with ATP and L-lysine
To be Published
|
|
7BAN
 
 | | human Teneurin4 Mut C2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Meijer, D.H, Janssen, B.J.C. | | Deposit date: | 2020-12-16 | | Release date: | 2022-01-12 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Teneurin4 dimer structures reveal a calcium-stabilized compact conformation supporting homomeric trans-interactions.
Embo J., 41, 2022
|
|
7Y7P
 
 | | QDE-1 in complex with RNA template, RNA primer and AMPNPP | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cui, R.X, Gan, J.H, Ma, J.B. | | Deposit date: | 2022-06-22 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the dual activities of the two-barrel RNA polymerase QDE-1.
Nucleic Acids Res., 50, 2022
|
|
7Y7S
 
 | | QDE-1 in complex with DNA template, RNA primer and AMPNPP | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cui, R.X, Gan, J.H, Ma, J.B. | | Deposit date: | 2022-06-22 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the dual activities of the two-barrel RNA polymerase QDE-1.
Nucleic Acids Res., 50, 2022
|
|
5AUJ
 
 | |
7RL1
 
 | | AAVrh.10-7x capsid | | Descriptor: | Capsid protein VP1, DNA (5'-D(*CP*A)-3') | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural Study of Aavrh.10 Receptor and Antibody Interactions.
J.Virol., 95, 2021
|
|
5FJ8
 
 | | Cryo-EM structure of yeast RNA polymerase III elongation complex at 3. 9 A | | Descriptor: | DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC1, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC10, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC2, ... | | Authors: | Hoffmann, N.A, Jakobi, A.J, Moreno-Morcillo, M, Glatt, S, Kosinski, J, Hagen, W.J, Sachse, C, Muller, C.W. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-25 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular Structures of Unbound and Transcribing RNA Polymerase III.
Nature, 528, 2015
|
|
3TWI
 
 | | Variable Lymphocyte Receptor Recognition of the Immunodominant Glycoprotein of Bacillus anthracis Spores | | Descriptor: | BclA protein, GLYCEROL, Variable lymphocyte receptor B | | Authors: | Kirchdoerfer, R.N, Herrin, B.R, Han, B.W, Turnbough Jr, C.L, Cooper, M.D, Wilson, I.A. | | Deposit date: | 2011-09-21 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Variable Lymphocyte Receptor Recognition of the Immunodominant Glycoprotein of Bacillus anthracis Spores.
Structure, 20, 2012
|
|
7R5M
 
 | |
7S1W
 
 | | The AAVrh.10-glycan complex | | Descriptor: | Capsid protein VP1, beta-D-galactopyranose | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2021-09-02 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural Study of Aavrh.10 Receptor and Antibody Interactions.
J.Virol., 95, 2021
|
|
5CJ2
 
 | | Ran GDP Y39A mutant triclinic crystal form | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Vetter, I.R, Brucker, S. | | Deposit date: | 2015-07-13 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Catalysis of GTP Hydrolysis by Small GTPases at Atomic Detail by Integration of X-ray Crystallography, Experimental, and Theoretical IR Spectroscopy.
J.Biol.Chem., 290, 2015
|
|
8RQB
 
 | | Cryo-EM structure of mouse heavy-chain apoferritin | | Descriptor: | FE (III) ION, Ferritin heavy chain, N-terminally processed, ... | | Authors: | Nazarov, S, Myasnikov, A, Stahlberg, H. | | Deposit date: | 2024-01-17 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (1.09 Å) | | Cite: | Low-dose cryo-electron ptychography of proteins at sub-nanometer resolution.
Nat Commun, 15, 2024
|
|
