2YYX
 
 | |
2IDT
 
 | | Structure of M98Q mutant of amicyanin, Cu(II) | | Descriptor: | Amicyanin, COPPER (II) ION | | Authors: | Carrell, C.J, Ma, J.K, Antholine, W, Hosler, J.P, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2006-09-15 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Generation of Novel Copper Sites by Mutation of the Axial Ligand of Amicyanin. Atomic Resolution Structures and Spectroscopic Properties
Biochemistry, 46, 2007
|
|
2P5K
 
 | | Crystal structure of the N-terminal domain of AhrC | | Descriptor: | Arginine repressor | | Authors: | Garnett, J.A, Baumberg, S, Stockley, P.G, Phillips, S.E.V. | | Deposit date: | 2007-03-15 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A high-resolution structure of the DNA-binding domain of AhrC, the arginine repressor/activator protein from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2X46
 
 | | Crystal Structure of SeMet Arg r 1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALLERGEN ARG R 1 | | Authors: | Paesen, G.C, Siebold, C, Syme, N, Harlos, K, Graham, S.C, Hilger, C, Homans, S.W, Hentges, F, Stuart, D.I. | | Deposit date: | 2010-01-28 | | Release date: | 2011-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal Structure of the Allergen Arg R 1, a Histamine-Binding Lipocalin from a Soft Tick
To be Published
|
|
8C10
 
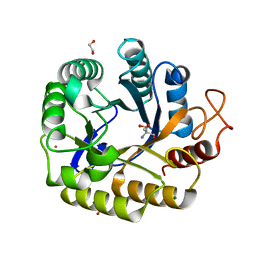 | | Biochemical and structural characterisation of an alkaline family GH5 cellulase from a shipworm symbiont | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GH5 Cellulase, ... | | Authors: | Leiros, I, Vaaje-Kolstad, G. | | Deposit date: | 2022-12-19 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Biochemical and structural characterisation of a family GH5 cellulase from endosymbiont of shipworm P. megotara.
Biotechnol Biofuels Bioprod, 16, 2023
|
|
1GKM
 
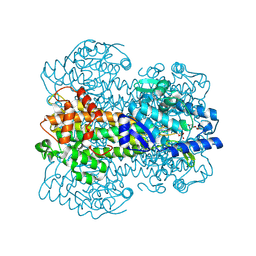 | |
1T2H
 
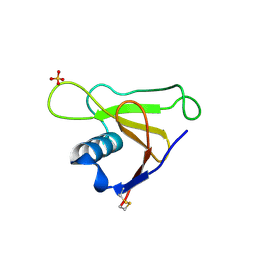 | |
4LN2
 
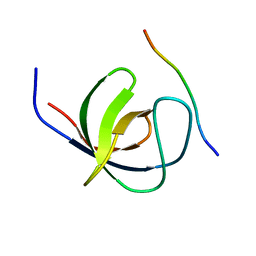 | | The second SH3 domain from CAP/Ponsin in complex with proline rich peptide from Vinculin | | Descriptor: | Sorbin and SH3 domain-containing protein 1, proline rich peptide | | Authors: | Zhao, D, Li, F, Wu, J, Shi, Y, Zhang, Z, Gong, Q. | | Deposit date: | 2013-07-11 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural investigation of the interaction between the tandem SH3 domains of c-Cbl-associated protein and vinculin
J.Struct.Biol., 187, 2014
|
|
2CE2
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A FLUORESCENT FORM OF H-RAS P21 IN COMPLEX WITH GDP | | Descriptor: | GTPASE HRAS, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Klink, B.U, Goody, R.S, Scheidig, A.J. | | Deposit date: | 2006-02-02 | | Release date: | 2006-08-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A Newly Designed Microspectrofluorometer for Kinetic Studies on Protein Crystals in Combination with X-Ray Diffraction
Biophys.J., 91, 2006
|
|
8B8F
 
 | | Atomic structure of the beta-trefoil domain of the Laccaria bicolor lectin LBL in complex with lactose | | Descriptor: | N-terminal beta-trefoil domain of the lectin LBL from Laccaria bicolor, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Acebron, I, Campanero-Rhodes, M.A, Solis, D, Menendez, M, Garcia, C, Lillo, M.P, Mancheno, J.M. | | Deposit date: | 2022-10-04 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic crystal structure and sugar specificity of a beta-trefoil lectin domain from the ectomycorrhizal basidiomycete Laccaria bicolor.
Int.J.Biol.Macromol., 233, 2023
|
|
1Y55
 
 | | Crystal structure of the C122S mutant of E. Coli expressed avidin related protein 4 (AVR4)-biotin complex | | Descriptor: | Avidin-related protein 4/5, BIOTIN, FORMIC ACID | | Authors: | Eisenberg-Domovich, Y, Hytonen, V.P, Wilchek, M, Bayer, E.A, Kulomaa, M.S, Livnah, O. | | Deposit date: | 2004-12-02 | | Release date: | 2005-05-24 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution crystal structure of an avidin-related protein: insight into high-affinity biotin binding and protein stability.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4GNR
 
 | | 1.0 Angstrom resolution crystal structure of the branched-chain amino acid transporter substrate binding protein LivJ from Streptococcus pneumoniae str. Canada MDR_19A in complex with Isoleucine | | Descriptor: | ABC transporter substrate-binding protein-branched chain amino acid transport, CHLORIDE ION, ISOLEUCINE, ... | | Authors: | Halavaty, A.S, Kudritska, M, Wawrzak, Z, Stogios, P.J, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-17 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | 1.0 Angstrom resolution crystal structure of the branched-chain amino acid transporter substrate binding protein LivJ from Streptococcus pneumoniae str. Canada MDR_19A in complex with Isoleucine
To be Published
|
|
6Q49
 
 | | CDK2 in complex with FragLite6 | | Descriptor: | 4-bromanyl-1~{H}-pyridin-2-one, Cyclin-dependent kinase 2, DIMETHYL SULFOXIDE | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
1A6M
 
 | | OXY-MYOGLOBIN, ATOMIC RESOLUTION | | Descriptor: | MYOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Vojtechovsky, J, Chu, K, Berendzen, J, Sweet, R.M, Schlichting, I. | | Deposit date: | 1998-02-26 | | Release date: | 1999-04-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal structures of myoglobin-ligand complexes at near-atomic resolution.
Biophys.J., 77, 1999
|
|
8DA5
 
 | | Coevolved affibody-Z domain pair LL1.c4 | | Descriptor: | GLYCEROL, Immunoglobulin G-binding protein A, affibody LL1.FIVM | | Authors: | Jude, K.M, Yang, A, Garcia, K.C. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Deploying synthetic coevolution and machine learning to engineer protein-protein interactions.
Science, 381, 2023
|
|
6NFR
 
 | | CopC from Pseudomonas fluorescens | | Descriptor: | CopC, SULFATE ION | | Authors: | Maher, M.J. | | Deposit date: | 2018-12-20 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The crystal structure of the CopC protein from Pseudomonas fluorescens reveals amended classifications for the CopC protein family.
J. Inorg. Biochem., 195, 2019
|
|
8JFS
 
 | | Phosphate bound acylphosphatase from Deinococcus radiodurans at 1 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, Acylphosphatase, CITRIC ACID, ... | | Authors: | Khakerwala, Z, Kumar, A, Makde, R.D. | | Deposit date: | 2023-05-18 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal structure of phosphate bound Acyl phosphatase mini-enzyme from Deinococcus radiodurans at 1 angstrom resolution.
Biochem.Biophys.Res.Commun., 671, 2023
|
|
1C7K
 
 | | CRYSTAL STRUCTURE OF THE ZINC PROTEASE | | Descriptor: | CALCIUM ION, ZINC ENDOPROTEASE, ZINC ION | | Authors: | Kurisu, G, Harada, S, Kai, Y. | | Deposit date: | 2000-02-19 | | Release date: | 2001-04-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure of the zinc-binding site in the crystal structure of a zinc endoprotease from Streptomyces caespitosus at 1 A resolution.
J.Inorg.Biochem., 82, 2000
|
|
5JIG
 
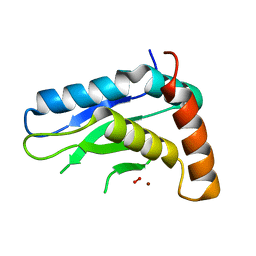 | | Crytsal structure of Wss1 from S. pombe | | Descriptor: | NICKEL (II) ION, OXYGEN MOLECULE, Ubiquitin and WLM domain-containing metalloprotease SPCC1442.07c | | Authors: | Groll, M, Stingele, J, Boulton, S. | | Deposit date: | 2016-04-22 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Mechanism and Regulation of DNA-Protein Crosslink Repair by the DNA-Dependent Metalloprotease SPRTN.
Mol.Cell, 64, 2016
|
|
2OFR
 
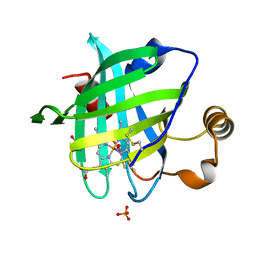 | |
3NE0
 
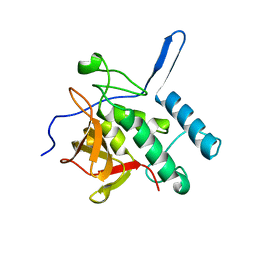 | |
6Q4H
 
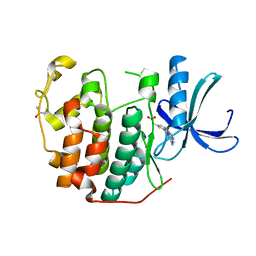 | | CDK2 in complex with FragLite36 | | Descriptor: | 2-[3-[(2-azanyl-9~{H}-purin-6-yl)oxy]phenyl]ethanoic acid, Cyclin-dependent kinase 2, DIMETHYL SULFOXIDE | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
7TMJ
 
 | |
2V1M
 
 | | Crystal structure of Schistosoma mansoni glutathione peroxidase | | Descriptor: | ACETATE ION, GLUTATHIONE PEROXIDASE, LITHIUM ION, ... | | Authors: | Dimastrogiovanni, D, Miele, A.E, Angelucci, F, Boumis, G, Bellelli, A, Brunori, M. | | Deposit date: | 2008-09-16 | | Release date: | 2009-09-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Combining Crystallography and Molecular Dynamics: The Case of Schistosoma Mansoni Phospholipid Glutathione Peroxidase.
Proteins, 78, 2010
|
|
1UZV
 
 | | High affinity fucose binding of Pseudomonas aeruginosa lectin II: 1.0 A crystal structure of the complex | | Descriptor: | CALCIUM ION, PSEUDOMONAS AERUGINOSA LECTIN II, SULFATE ION, ... | | Authors: | Mitchell, E, Sabin, C.D, Snajdrova, L, Budova, M, Perret, S, Gautier, C, Gilboa-Garber, N, Koca, J, Wimmerova, M, Imberty, A. | | Deposit date: | 2004-03-17 | | Release date: | 2004-12-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High Affinity Fucose Binding of Pseudomonas Aeruginosa Lectin Pa-Iil: 1.0 A Resolution Crystal Structure of the Complex Combined with Thermodynamics and Computational Chemistry Approaches.
Proteins, 58, 2005
|
|
