5J6M
 
 | | Crystal Structure of Hsp90-alpha N-domain L107 mutant in complex with 5-[4-(2-Fluoro-phenyl)-5-oxo-4,5-dihydro-1H-[1,2,4]triazol-3-yl]-N-furan-2-ylmethyl-2,4-dihydroxy-N-methyl-benzamide | | Descriptor: | 5-[4-(2-fluorophenyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl]-N-[(furan-2-yl)methyl]-2,4-dihydroxy-N-methylbenzamide, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-05 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
5J8M
 
 | | Crystal Structure of Hsp90-alpha N-domain L107A mutant in complex with 5-(5-Bromo-2,4-dihydroxy-phenyl)-4-(2-fluoro-phenyl)-2,4-dihydro-[1,2,4]triazol-3-one | | Descriptor: | 5-(5-Bromo-2,4-dihydroxy-phenyl)-4-(2-fluoro-phenyl)-2,4-dihydro-[1,2,4]triazol-3-one, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-08 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
7NUS
 
 | |
5G15
 
 | | Structure Aurora A (122-403) bound to activating monobody Mb1 and AMPPCP | | Descriptor: | AURORA A KINASE, MAGNESIUM ION, MB1 MONOBODY, ... | | Authors: | Zorba, A, Kutter, S, Kern, D, Koide, S, Koide, A. | | Deposit date: | 2016-03-23 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Allosteric modulation of a human protein kinase with monobodies.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5J9L
 
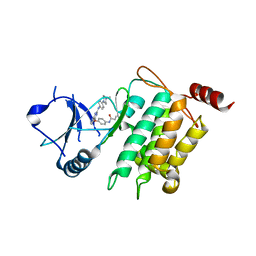 | | Crystal structure of CPT1691 bound to TAK1-TAB1 | | Descriptor: | Mitogen-activated protein kinase kinase kinase 7,TGF-beta-activated kinase 1 and MAP3K7-binding protein 1, N-(4-((2-((4-(4-methylpiperazin-1-yl)phenyl)amino)-7H-pyrrolo[2,3-d]pyrimidin-4-yl)oxy)phenyl)acrylamide | | Authors: | Gurbani, D, Westover, K.D. | | Deposit date: | 2016-04-10 | | Release date: | 2017-02-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7515 Å) | | Cite: | Structure-guided development of covalent TAK1 inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
5J9Z
 
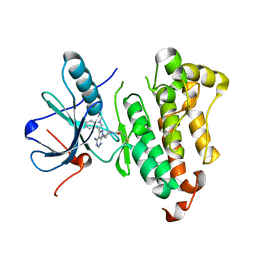 | | EGFR-T790M in complex with pyrazolopyrimidine inhibitor 1a | | Descriptor: | (R)-1-(3-(4-amino-3-(1-methyl-1H-indol-3-yl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl)piperidin-1-yl)prop-2-en-1-one, Epidermal growth factor receptor | | Authors: | Becker, C, Engel, J, Rauh, D. | | Deposit date: | 2016-04-11 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into the Inhibition of Drug-Resistant Mutants of the Receptor Tyrosine Kinase EGFR.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
7OJ7
 
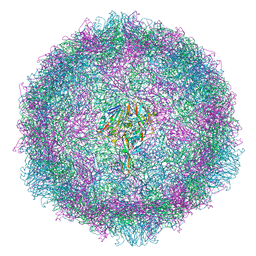 | |
7O2R
 
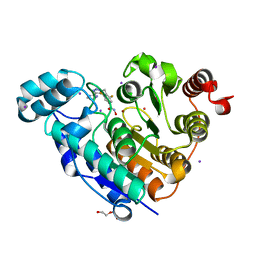 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 in complex with ITF3985 | | Descriptor: | 3,5-bis(fluoranyl)-~{N}-oxidanyl-4-[(5-pyrimidin-2-yl-1,2,3,4-tetrazol-2-yl)methyl]benzamide, DI(HYDROXYETHYL)ETHER, Histone deacetylase 6, ... | | Authors: | Zrubek, K, Sandrone, G, Cukier, C.D, Stevenazzi, A. | | Deposit date: | 2021-03-31 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of Fluorination in the Histone Deacetylase 6 (HDAC6) Selectivity of Benzohydroxamate-Based Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
7O2P
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 in complex with ITF3756 | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zrubek, K, Sandrone, G, Cukier, C.D, Stevenazzi, A. | | Deposit date: | 2021-03-31 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of Fluorination in the Histone Deacetylase 6 (HDAC6) Selectivity of Benzohydroxamate-Based Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
5G0T
 
 | | InhA in complex with a DNA encoded library hit | | Descriptor: | 1-benzyl-N-[cis-4-(2-{[(4-fluorophenyl)methyl][2-(methylamino)-2-oxoethyl]amino}-2-oxoethyl)cyclohexyl]-5-methyl-1H-1,2,3-triazole-4-carboxamide, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Read, J.A, Breed, J. | | Deposit date: | 2016-03-22 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Discovery of Cofactor-Specific, Bactericidal Mycobacterium Tuberculosis Inha Inhibitors Using DNA-Encoded Library Technology
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5J1H
 
 | |
5J1I
 
 | |
7NSL
 
 | |
7O90
 
 | | Mono-Fe-sulerythrin | | Descriptor: | CHLORIDE ION, FE (III) ION, Sulerythrin | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2021-04-15 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Bimetallic Mn, Fe, Co, and Ni Sites in a Four-Helix Bundle Protein: Metal Binding, Structure, and Peroxide Activation.
Inorg.Chem., 60, 2021
|
|
7O93
 
 | | diMn-sulerythrin | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CHLORIDE ION, HYDROGEN PEROXIDE, ... | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2021-04-15 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Bimetallic Mn, Fe, Co, and Ni Sites in a Four-Helix Bundle Protein: Metal Binding, Structure, and Peroxide Activation.
Inorg.Chem., 60, 2021
|
|
7O8D
 
 | | diFe-sulerythrin oxidised by H2O2 | | Descriptor: | CHLORIDE ION, FE (III) ION, Sulerythrin | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2021-04-15 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Bimetallic Mn, Fe, Co, and Ni Sites in a Four-Helix Bundle Protein: Metal Binding, Structure, and Peroxide Activation.
Inorg.Chem., 60, 2021
|
|
7O9D
 
 | | peroxide-bound diCo-sulerythrin | | Descriptor: | CHLORIDE ION, COBALT (II) ION, HYDROGEN PEROXIDE, ... | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2021-04-15 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Bimetallic Mn, Fe, Co, and Ni Sites in a Four-Helix Bundle Protein: Metal Binding, Structure, and Peroxide Activation.
Inorg.Chem., 60, 2021
|
|
7O9C
 
 | | diNi-sulerythrin treated by hydrogen perxoide | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Sulerythrin | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2021-04-15 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bimetallic Mn, Fe, Co, and Ni Sites in a Four-Helix Bundle Protein: Metal Binding, Structure, and Peroxide Activation.
Inorg.Chem., 60, 2021
|
|
7O89
 
 | | sulerythrin without metals (apo-state) | | Descriptor: | CHLORIDE ION, Sulerythrin | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2021-04-15 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Bimetallic Mn, Fe, Co, and Ni Sites in a Four-Helix Bundle Protein: Metal Binding, Structure, and Peroxide Activation.
Inorg.Chem., 60, 2021
|
|
7O9E
 
 | | diNi-sulerythrin | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Sulerythrin | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2021-04-15 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Bimetallic Mn, Fe, Co, and Ni Sites in a Four-Helix Bundle Protein: Metal Binding, Structure, and Peroxide Activation.
Inorg.Chem., 60, 2021
|
|
5J7B
 
 | | The identification and pharmacological characterization of 6-(tert-butylsulfonyl)-N-(5-fluoro-1H-indazol-3-yl)quinolin-4-amine (GSK583), a highly potent and selective inhibitor of RIP2 Kinase, GSK583 complex | | Descriptor: | 6-(tert-butylsulfonyl)-N-(5-fluoro-2H-indazol-3-yl)quinolin-4-amine, Receptor-interacting serine/threonine-protein kinase 2 | | Authors: | Convery, M.A, Casillas, L.N, Haile, P.A, Votta, B.J, Lakdawala, A.S. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The Identification and Pharmacological Characterization of 6-(tert-Butylsulfonyl)-N-(5-fluoro-1H-indazol-3-yl)quinolin-4-amine (GSK583), a Highly Potent and Selective Inhibitor of RIP2 Kinase.
J.Med.Chem., 59, 2016
|
|
7O8A
 
 | | diFe-sulerythrin reduced with Na-dithionite | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, FE (III) ION, ... | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2021-04-15 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Bimetallic Mn, Fe, Co, and Ni Sites in a Four-Helix Bundle Protein: Metal Binding, Structure, and Peroxide Activation.
Inorg.Chem., 60, 2021
|
|
7O99
 
 | | diMn-sulerythrin | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Sulerythrin | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2021-04-15 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Bimetallic Mn, Fe, Co, and Ni Sites in a Four-Helix Bundle Protein: Metal Binding, Structure, and Peroxide Activation.
Inorg.Chem., 60, 2021
|
|
5J80
 
 | |
5J83
 
 | |
