4YNT
 
 | | Crystal structure of Aspergillus flavus FAD glucose dehydrogenase | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Glucose oxidase, putative | | Authors: | Yoshida, H, Sakai, G, Kojima, K, Kamitori, S, Sode, K. | | Deposit date: | 2015-03-11 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural analysis of fungus-derived FAD glucose dehydrogenase
Sci Rep, 5, 2015
|
|
4O9U
 
 | | Mechanism of transhydrogenase coupling proton translocation and hydride transfer | | Descriptor: | NAD(P) transhydrogenase subunit alpha 2, NAD(P) transhydrogenase subunit beta, NAD/NADP transhydrogenase alpha subunit 1, ... | | Authors: | Leung, J.H, Yamaguchi, M, Moeller, A, Schurig-Briccio, L.A, Gennis, R.B, Potter, C.S, Carragher, B, Stout, C.D. | | Deposit date: | 2014-01-02 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (6.926 Å) | | Cite: | Structural biology. Division of labor in transhydrogenase by alternating proton translocation and hydride transfer.
Science, 347, 2015
|
|
4OAP
 
 | | An Axe2 mutant (W190I), an acetyl-xylooligosaccharide esterase from Geobacillus Stearmophilus | | Descriptor: | ACETATE ION, Acetyl xylan esterase, CHLORIDE ION, ... | | Authors: | Lansky, S, Alalouf, O, Solomon, H.V, Belrhali, H, Shoham, Y, Shoham, G. | | Deposit date: | 2014-01-06 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | An Axe2 mutant (W190I), an acetyl-xylooligosaccharide esterase from Geobacillus Stearmophilus
To be Published
|
|
4YNA
 
 | | Oxidized YfiR | | Descriptor: | SULFATE ION, YfiR | | Authors: | Xu, M, Jiang, T. | | Deposit date: | 2015-03-09 | | Release date: | 2015-04-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of YfiR from Pseudomonas aeruginosa in two redox states
Biochem.Biophys.Res.Commun., 461, 2015
|
|
4OWF
 
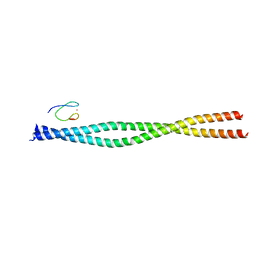 | | Crystal structure of the NEMO CoZi in complex with HOIP NZF1 domain | | Descriptor: | E3 ubiquitin-protein ligase RNF31, NF-kappa-B essential modulator, ZINC ION | | Authors: | Rahighi, S, Fujita, H, Kawasaki, M, Kato, R, Iwai, K, Wakatsuki, S. | | Deposit date: | 2014-01-31 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism Underlying I kappa B Kinase Activation Mediated by the Linear Ubiquitin Chain Assembly Complex.
Mol.Cell.Biol., 34, 2014
|
|
4YP7
 
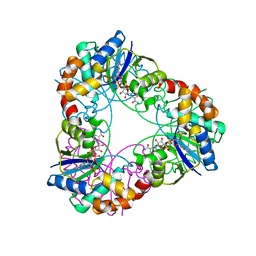 | | Crystal structure of Methanobacterium thermoautotrophicum NMNAT in complex with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Nicotinamide-nucleotide adenylyltransferase | | Authors: | Pfoh, R, Christendat, D, Pai, E.F, Saridakis, V. | | Deposit date: | 2015-03-12 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nicotinamide mononucleotide adenylyltransferase displays alternate binding modes for nicotinamide nucleotides.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
4OBJ
 
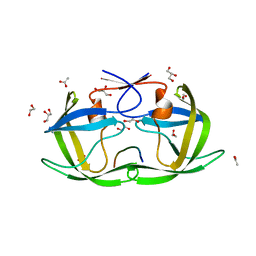 | |
4OWQ
 
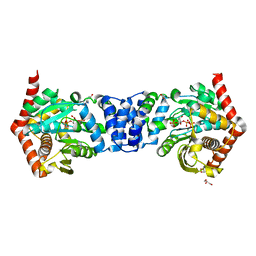 | | Anthranilate phosphoribosyl transferase from Mycobacterium tuberculosis in complex with 3-methylanthranilate, PRPP and Magnesium | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 2-azanyl-3-methyl-benzoic acid, Anthranilate phosphoribosyltransferase, ... | | Authors: | Castell, A, Cookson, T.V.M, Short, F.L, Lott, J.S. | | Deposit date: | 2014-02-03 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Alternative substrates reveal catalytic cycle and key binding events in the reaction catalysed by anthranilate phosphoribosyltransferase from Mycobacterium tuberculosis.
Biochem.J., 461, 2014
|
|
4OBW
 
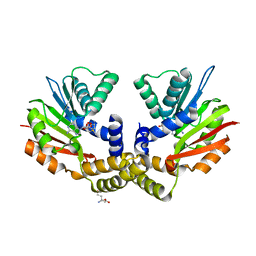 | | crystal structure of yeast Coq5 in the SAM bound form | | Descriptor: | 2-methoxy-6-polyprenyl-1,4-benzoquinol methylase, mitochondrial, S-ADENOSYLMETHIONINE, ... | | Authors: | Dai, Y.N, Zhou, K, Cao, D.D, Jiang, Y.L, Meng, F, Chi, C.B, Ren, Y.M, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2014-01-07 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures and catalytic mechanism of the C-methyltransferase Coq5 provide insights into a key step of the yeast coenzyme Q synthesis pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4YPU
 
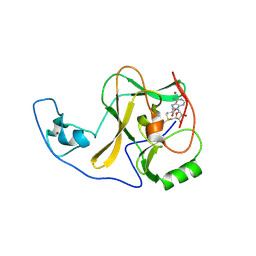 | | ASH1L SET domain K2264L mutant in complex with S-adenosyl methionine (SAM) | | Descriptor: | Histone-lysine N-methyltransferase ASH1L, S-ADENOSYLMETHIONINE, ZINC ION | | Authors: | Rogawski, D.S, Ndoj, J, Cho, H.J, Maillard, I, Grembecka, J, Cierpicki, T. | | Deposit date: | 2015-03-13 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Two Loops Undergoing Concerted Dynamics Regulate the Activity of the ASH1L Histone Methyltransferase.
Biochemistry, 54, 2015
|
|
4OCJ
 
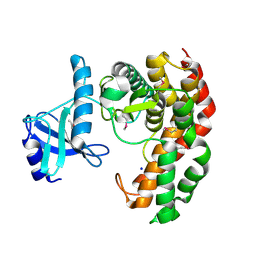 | | N-acetylhexosamine 1-phosphate kinase in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, N-acetylhexosamine 1-phosphate kinase, SODIUM ION | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OCU
 
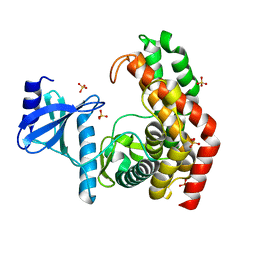 | | N-acetylhexosamine 1-phosphate kinase_ATCC15697 in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, N-acetylhexosamine 1-phosphate kinase, SULFATE ION | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OYL
 
 | |
4OD0
 
 | | Crystal structure of human soluble epoxide hydrolase complexed with 1-(1-propanoylpiperidin-4-yl)-3-[4-(trifluoromethoxy)phenyl]urea | | Descriptor: | 1-(1-propanoylpiperidin-4-yl)-3-[4-(trifluoromethoxy)phenyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Lee, K.S.S, Liu, J, Wagner, K.M, Pakhomova, S, Dong, H, Morisseau, C, Fu, S.H, Yang, J, Wang, P, Ulu, A, Mate, C, Nguyen, L, Wullf, H, Eldin, M.L, Mara, A.A, Newcomer, M.E, Zeldin, D.C, Hammock, B.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Optimized inhibitors of soluble epoxide hydrolase improve in vitro target residence time and in vivo efficacy.
J.Med.Chem., 57, 2014
|
|
4ODG
 
 | | Crystal structure of Staphylococcal nuclease variant V23I/V66I/V74I/V99I at cryogenic temperature | | Descriptor: | CALCIUM ION, PHOSPHATE ION, THYMIDINE-3',5'-DIPHOSPHATE, ... | | Authors: | Caro, J.A, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2014-01-10 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
4OJP
 
 | |
4ODW
 
 | | Unliganded Fab structure of lipid A-specific antibody A6 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, A6 Fab (IgG2b kappa) light chain, A6 Fab (IgG2b) heavy chain | | Authors: | Haji-Ghassemi, O, Evans, S.V. | | Deposit date: | 2014-01-10 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural Basis for Antibody Recognition of Lipid A: INSIGHTS TO POLYSPECIFICITY TOWARD SINGLE-STRANDED DNA.
J.Biol.Chem., 290, 2015
|
|
4YR9
 
 | | mouse TDH with NAD+ bound | | Descriptor: | GLYCEROL, L-threonine 3-dehydrogenase, mitochondrial, ... | | Authors: | He, C, Li, F. | | Deposit date: | 2015-03-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights on mouse l-threonine dehydrogenase: A regulatory role of Arg180 in catalysis
J.Struct.Biol., 192, 2015
|
|
4OFT
 
 | | C- Orthorombic NaGST1 | | Descriptor: | Glutathione S-transferase-1 | | Authors: | Asojo, O.A. | | Deposit date: | 2014-01-15 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of glutathione S-transferase 1 from the major human hookworm parasite Necator americanus (Na-GST-1) in complex with glutathione.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4YSU
 
 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 25.0 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4OG4
 
 | | Human menin with bound inhibitor MIV-3S | | Descriptor: | 4-(3-{4-[(S)-cyclopentyl(hydroxy)phenylmethyl]piperidin-1-yl}propoxy)benzonitrile, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | He, S, Senter, T.J, Pollock, J.W, Han, C, Upadhyay, S.K, Purohit, T, Gogliotti, R.D, Lindsley, C.W, Cierpicki, T, Stauffer, S.R, Grembecka, J. | | Deposit date: | 2014-01-15 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-Affinity Small-Molecule Inhibitors of the Menin-Mixed Lineage Leukemia (MLL) Interaction Closely Mimic a Natural Protein-Protein Interaction.
J.Med.Chem., 57, 2014
|
|
4OGI
 
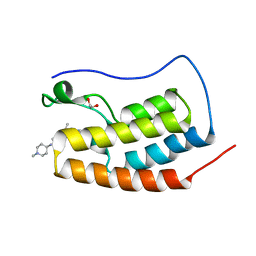 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor BI-2536 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Jose, B, Martin, S, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-01-16 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Dual kinase-bromodomain inhibitors for rationally designed polypharmacology.
Nat.Chem.Biol., 10, 2014
|
|
4YSH
 
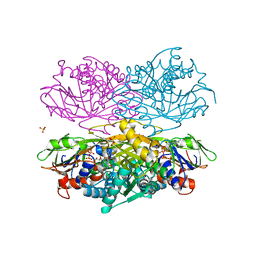 | |
4OGS
 
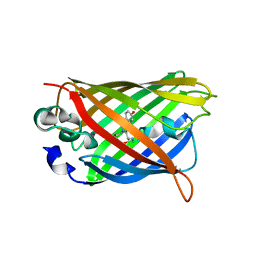 | |
4YSZ
 
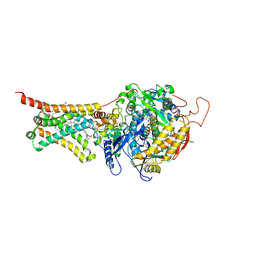 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with 2-iodo-N-[3-(1-methylethoxy)phenyl]benzamide | | Descriptor: | 2-iodo-N-[3-(1-methylethoxy)phenyl]benzamide, Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-03-17 | | Release date: | 2015-08-05 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
