7AGD
 
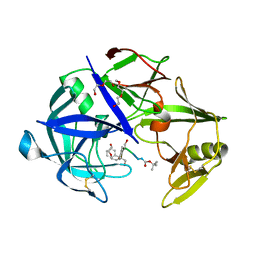 | | Protease Sapp1p from Candida parapsilosis in complex with KB75 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Candidapepsin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dostal, J, Heidingsfeld, O, Brynda, J. | | Deposit date: | 2020-09-22 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural determinants for subnanomolar inhibition of the secreted aspartic protease Sapp1p from Candida parapsilosis .
J Enzyme Inhib Med Chem, 36, 2021
|
|
7AGC
 
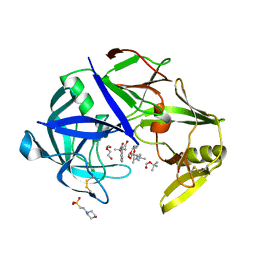 | | Protease Sapp1p from Candida parapsilosis in complex with KB74 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Candidapepsin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dostal, J, Heidingsfeld, O, Brynda, J. | | Deposit date: | 2020-09-22 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural determinants for subnanomolar inhibition of the secreted aspartic protease Sapp1p from Candida parapsilosis .
J Enzyme Inhib Med Chem, 36, 2021
|
|
7B1Q
 
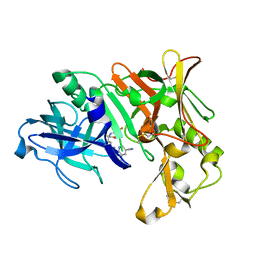 | | Crystal Structure of Human BACE-1 in Complex with Compound NB-360 (compound 54) | | Descriptor: | Beta-secretase 1, ~{N}-[3-[(3~{R},6~{R})-5-azanyl-3,6-dimethyl-6-(trifluoromethyl)-2~{H}-1,4-oxazin-3-yl]-4-fluoranyl-phenyl]-5-cyano-3-methyl-pyridine-2-carboxamide | | Authors: | Rondeau, J.M, Wirth, E. | | Deposit date: | 2020-11-25 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Synthesis of the Potent, Selective, and Efficacious beta-Secretase (BACE1) Inhibitor NB-360.
J.Med.Chem., 64, 2021
|
|
7B1E
 
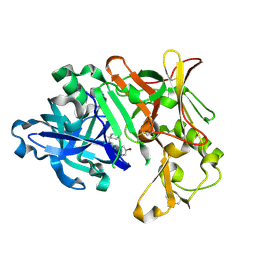 | | BACE1 IN COMPLEX WITH compound 3 (NB-641) | | Descriptor: | Beta-secretase 1, ~{N}-[3-[(4~{S})-2-azanyl-4-methyl-5,6-dihydro-1,3-thiazin-4-yl]phenyl]-5-bromanyl-pyridine-2-carboxamide | | Authors: | Rondeau, J.M, Wirth, E. | | Deposit date: | 2020-11-24 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Synthesis of the Potent, Selective, and Efficacious beta-Secretase (BACE1) Inhibitor NB-360.
J.Med.Chem., 64, 2021
|
|
7B1P
 
 | | Crystal Structure of Human BACE-1 in Complex with Compound 38a (NB-854) | | Descriptor: | Beta-secretase 1, ~{N}-[3-[(3~{R},6~{R})-5-azanyl-3,6-dimethyl-6-(trifluoromethyl)-2~{H}-1,4-oxazin-3-yl]phenyl]-5-bromanyl-pyridine-2-carboxamide | | Authors: | Rondeau, J.M, Wirth, E. | | Deposit date: | 2020-11-25 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Synthesis of the Potent, Selective, and Efficacious beta-Secretase (BACE1) Inhibitor NB-360.
J.Med.Chem., 64, 2021
|
|
8CIK
 
 | | Altai wapiti (Cervus elaphus sibiricus) chymosin at 2.2 A resolution | | Descriptor: | Chymosin, GLYCEROL | | Authors: | Diusenova, S.E, Shevtsov, M.B, Borshchevskiy, V.I, Belenkaya, S.V, Kolybalov, D.S, Arkhipov, S.G, Volosnikova, E.A, Elchaninov, V.V, Shcherbakov, D.N. | | Deposit date: | 2023-02-09 | | Release date: | 2023-03-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Altai wapiti (Cervus elaphus sibiricus) chymosin at 2.2 A resolution
To Be Published
|
|
6JSG
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(4S)-2-amino-4-methyl-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-chloropyridine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
6JSZ
 
 | | BACE2 xaperone complex with N-{3-[(5R)-3-amino-5-methyl-9,9-dioxo-2,9lambda6-dithia-4-azaspiro[5.5]undec-3-en-5-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 2, CHLORIDE ION, N-[3-[(5R)-3-azanyl-5-methyl-9,9-bis(oxidanylidene)-2,9$l^{6}-dithia-4-azaspiro[5.5]undec-3-en-5-yl]-4-fluoranyl-phenyl]-5-(fluoranylmethoxy)pyrazine-2-carboxamide, ... | | Authors: | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
6JSN
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(5R)-3-amino-5-methyl-9,9-dioxo-2,9lambda6-dithia-4-azaspiro[5.5]undec-3-en-5-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
6JSF
 
 | | Crystal Structure of BACE1 in complex with N-(3-((4S,5S)-2-amino-4-methyl-5-phenyl-5,6-dihydro-4H-1,3-thiazin-4-yl)-4-fluorophenyl)-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, IODIDE ION, ... | | Authors: | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
6JT3
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(4R,5R,6R)-2-amino-5-fluoro-4,6-dimethyl-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tadano, G, Komano, K, Yoshida, S, Suzuki, S, Nakahara, K, Fuchino, K, Fujimoto, K, Matsuoka, E, Yamamoto, T, Asada, N, Ito, H, Sakaguchi, G, Kanegawa, N, Kido, Y, Ando, S, Fukushima, T, Teisman, A, Urmaliya, V, Dhuyvetter, D, Borghys, H, Bergh, A.V.D, Austin, N, Gijsen, H.J.M, Yamano, Y, Iso, Y, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-10-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of an Extremely Potent Thiazine-Based beta-Secretase Inhibitor with Reduced Cardiovascular and Liver Toxicity at a Low Projected Human Dose.
J.Med.Chem., 62, 2019
|
|
6JT4
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(4S,6S)-2-amino-4-methyl-6-(trifluoromethyl)-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Anan, K, Iso, Y, Oguma, T, Nakahara, K, Suzuki, S, Yamamoto, T, Matsuoka, E, Ito, H, Sakaguchi, G, Ando, S, Morimoto, K, Kanegawa, N, Kido, Y, Kawachi, T, Fukushima, T, Teisman, A, Urmaliya, V, Dhuyvetter, D, Borghys, H, Austin, N, Bergh, A.V.D, Verboven, P, Bischoff, F, Gijsen, H.J.M, Yamano, Y, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trifluoromethyl Dihydrothiazine-Based beta-Secretase (BACE1) Inhibitors with Robust Central beta-Amyloid Reduction and Minimal Covalent Binding Burden.
Chemmedchem, 14, 2019
|
|
6JSE
 
 | | Crystal Structure of BACE1 in complex with N-(3-((4S,5R)-2-amino-4-methyl-5-phenyl-5,6-dihydro-4H-1,3-thiazin-4-yl)-4-fluorophenyl)-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, IODIDE ION, ... | | Authors: | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
6KUC
 
 | | Crystal structure of Plasmodium falciparum histo-aspartic protease (HAP) zymogen (Form 2) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, HAP protein | | Authors: | Rathore, I, Mishra, V, Bhaumik, P. | | Deposit date: | 2019-08-31 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Activation mechanism of plasmepsins, pepsin-like aspartic proteases from Plasmodium, follows a unique trans-activation pathway.
Febs J., 288, 2021
|
|
6KUD
 
 | | Crystal structure of Plasmodium falciparum histo-aspartic protease (HAP) zymogen (Form 3) | | Descriptor: | GLYCEROL, HAP protein | | Authors: | Rathore, I, Mishra, V, Bhaumik, P. | | Deposit date: | 2019-08-31 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Activation mechanism of plasmepsins, pepsin-like aspartic proteases from Plasmodium, follows a unique trans-activation pathway.
Febs J., 288, 2021
|
|
6KUB
 
 | | Crystal structure of Plasmodium falciparum histo-aspartic protease (HAP) zymogen (Form 1) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, HAP protein | | Authors: | Rathore, I, Mishra, V, Bhaumik, P. | | Deposit date: | 2019-08-31 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Activation mechanism of plasmepsins, pepsin-like aspartic proteases from Plasmodium, follows a unique trans-activation pathway.
Febs J., 288, 2021
|
|
1IBQ
 
 | | ASPERGILLOPEPSIN FROM ASPERGILLUS PHOENICIS | | Descriptor: | ASPERGILLOPEPSIN, ZINC ION, alpha-D-mannopyranose | | Authors: | Cho, S.W, Shin, W. | | Deposit date: | 2001-03-28 | | Release date: | 2001-07-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure of aspergillopepsin I from Aspergillus phoenicis: variations of the S1'-S2 subsite in aspartic proteinases.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1YX9
 
 | |
1GVV
 
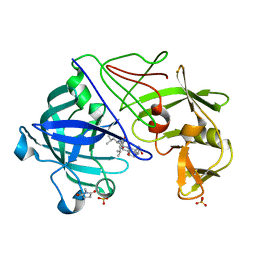 | | Five Atomic Resolution Structures of Endothiapepsin Inhibitor Complexes; implications for the Aspartic Proteinase Mechanism | | Descriptor: | ENDOTHIAPEPSIN, N-[(5S,9S,10S,13S)-9-hydroxy-5,10-bis(2-methylpropyl)-4,7,12,16-tetraoxo-3,6,11,17-tetraazabicyclo[17.3.1]tricosa-1(23),19,21-trien-13-yl]-3-(naphthalen-1-yl)-2-(naphthalen-1-ylmethyl)propanamide, SULFATE ION | | Authors: | Coates, L, Erskine, P.T, Crump, M.P, Wood, S.P, Cooper, J.B. | | Deposit date: | 2002-02-27 | | Release date: | 2002-07-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Five Atomic Resolution Structures of Endothiapepsin Inhibitor Complexes: Implications for the Aspartic Proteinase Mechanism
J.Mol.Biol., 318, 2002
|
|
1GVU
 
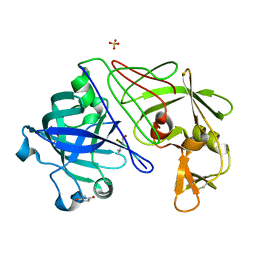 | | Endothiapepsin complex with H189 | | Descriptor: | ENDOTHIAPEPSIN, INHIBITOR, H189, ... | | Authors: | Coates, L, Erskine, P.T, Crump, M.P, Wood, S.P, Cooper, J.B. | | Deposit date: | 2002-02-27 | | Release date: | 2002-07-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Five Atomic Resolution Structures of Endothiapepsin Inhibitor Complexes: Implications for the Aspartic Proteinase Mechanism
J.Mol.Biol., 318, 2002
|
|
1YM4
 
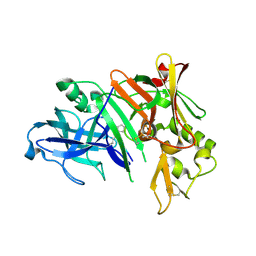 | | Crystal structure of human beta secretase complexed with NVP-AMK640 | | Descriptor: | Beta-secretase 1, NVP-AMK640 INHIBITOR | | Authors: | Hanessian, S, Yun, H, Hou, Y, Yang, G, Bayrakdarian, M, Therrien, E, Moitessier, N, Roggo, S, Veenstra, S. | | Deposit date: | 2005-01-20 | | Release date: | 2006-01-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based design, synthesis, and memapsin 2 (BACE) inhibitory activity of carbocyclic and heterocyclic peptidomimetics
J.Med.Chem., 48, 2005
|
|
2B8V
 
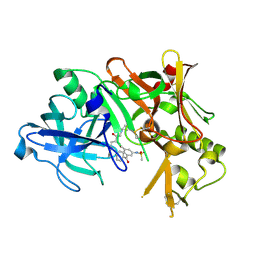 | | Crystal structure of human Beta-secretase complexed with L-L000430,469 | | Descriptor: | 3-BENZOYL-N-[(1S,2R)-1-BENZYL-3-(CYCLOPROPYLAMINO)-2-HYDROXYPROPYL]-5-[METHYL(METHYLSULFONYL)AMINO]BENZAMIDE, Beta-secretase 1 | | Authors: | Stachel, S.J, Coburn, C.A, Steele, T.G, Crouthamel, M.-C, Pietrak, B.L, Lai, M.-T, Holloway, M.K, Munshi, S.K, Graham, S.L, Vacca, J.P. | | Deposit date: | 2005-10-10 | | Release date: | 2005-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformationally biased P3 amide replacements of beta-secretase inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
8Q13
 
 | | Endothiapepsin in complex with ligand (3R,5R)-5-(3-(2-amino-4-(trifluoromethyl)phenyl)-1,2,4-oxadiazol-5-yl)-3-(2-((methyl(prop-2-yn-1-yl)amino)methyl)thiazol-4-yl)pyrrolidin-3-ol (CBWS-SE-168) | | Descriptor: | (3~{R},5~{R})-5-[3-[2-azanyl-4-(trifluoromethyl)phenyl]-1,2,4-oxadiazol-5-yl]-3-[2-[[methyl(prop-2-ynyl)amino]methyl]-1,3-thiazol-4-yl]pyrrolidin-3-ol, (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Mueller, J.M, Eckelt, S, Klebe, G, Glinca, S. | | Deposit date: | 2023-07-29 | | Release date: | 2024-08-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structures of Endothiapepsin with ligands derived from merged fragment hits
To Be Published
|
|
8Q11
 
 | | Endothiapepsin in complex with ligand (3R,5R)-5-(3-(4-fluorophenyl)-1,2,4-oxadiazol-5-yl)-3-(2-((methyl(prop-2-yn-1-yl)amino)methyl)thiazol-4-yl)pyrrolidin-3-ol (CBWS-SE-126) | | Descriptor: | (3~{R},5~{R})-5-[3-(4-fluorophenyl)-1,2,4-oxadiazol-5-yl]-3-[2-[[methyl(prop-2-ynyl)amino]methyl]-1,3-thiazol-4-yl]pyrrolidin-3-ol, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mueller, J.M, Eckelt, S, Klebe, G, Glinca, S. | | Deposit date: | 2023-07-29 | | Release date: | 2024-08-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of Endothiapepsin with ligands derived from merged fragment hits
To Be Published
|
|
1GVX
 
 | | Endothiapepsin complexed with H256 | | Descriptor: | ENDOTHIAPEPSIN, INHIBITOR H256, SULFATE ION | | Authors: | Coates, L, Erskine, P.T, Crump, M.P, Wood, S.P, Cooper, J.B. | | Deposit date: | 2002-02-27 | | Release date: | 2002-07-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Five Atomic Resolution Structures of Endothiapepsin Inhibitor Complexes: Implications for the Aspartic Proteinase Mechanism
J.Mol.Biol., 318, 2002
|
|
