3T2E
 
 | | Fructose-1,6-bisphosphate aldolase/phosphatase from Thermoproteus neutrophilus, F6P-bound form | | Descriptor: | FRUCTOSE -6-PHOSPHATE, Fructose-1,6-bisphosphate aldolase/phosphatase, MAGNESIUM ION | | Authors: | Du, J, Say, R, Lue, W, Fuchs, G, Einsle, O. | | Deposit date: | 2011-07-22 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Active-site remodelling in the bifunctional fructose-1,6-bisphosphate aldolase/phosphatase.
Nature, 478, 2011
|
|
3SUI
 
 | |
3SX2
 
 | |
3T6G
 
 | |
3T2B
 
 | | Fructose-1,6-bisphosphate aldolase/phosphatase from Thermoproteus neutrophilus, ligand free | | Descriptor: | Fructose-1,6-bisphosphate aldolase/phosphatase, MAGNESIUM ION | | Authors: | Du, J, Say, R, Lue, W, Fuchs, G, Einsle, O. | | Deposit date: | 2011-07-22 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Active-site remodelling in the bifunctional fructose-1,6-bisphosphate aldolase/phosphatase.
Nature, 478, 2011
|
|
3SJX
 
 | | X-ray structure of human glutamate carboxypeptidase II (the E424A inactive mutant) in complex with N-acetyl-aspartyl-methionine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Plechanovova, A, Byun, Y, Alquicer, G, Skultetyova, L, Mlcochova, P, Nemcova, A, Kim, H, Navratil, M, Mease, R, Lubkowski, J, Pomper, M, Konvalinka, J, Rulisek, L, Barinka, C. | | Deposit date: | 2011-06-22 | | Release date: | 2011-10-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Novel Substrate-Based Inhibitors of Human Glutamate Carboxypeptidase II with Enhanced Lipophilicity.
J.Med.Chem., 54, 2011
|
|
3SG2
 
 | | Crystal Structure of GCaMP2-T116V,D381Y | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Schreiter, E.R, Akerboom, J, Looger, L.L. | | Deposit date: | 2011-06-14 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Optimization of a GCaMP calcium indicator for neural activity imaging.
J.Neurosci., 32, 2012
|
|
3UT5
 
 | | Tubulin-Colchicine-Ustiloxin: Stathmin-like domain complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ranaivoson, F.M, Gigant, B, Knossow, M. | | Deposit date: | 2011-11-25 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structural plasticity of tubulin assembly probed by vinca-domain ligands.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3SOH
 
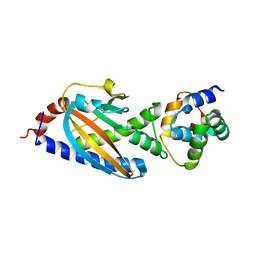 | | Architecture of the Flagellar Rotor | | Descriptor: | Flagellar motor switch protein FliG, Flagellar motor switch protein FliM | | Authors: | Koushik, P, Gonzalez-Bonet, G, Bilwes, A.M, Crane, B.R, Blair, D. | | Deposit date: | 2011-06-30 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Architecture of the flagellar rotor.
Embo J., 30, 2011
|
|
3SG6
 
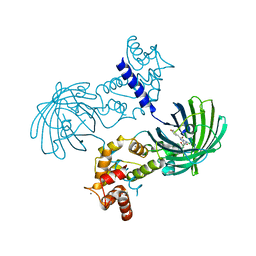 | | Crystal Structure of Dimeric GCaMP2-LIA(linker 1) | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Schreiter, E.R, Akerboom, J, Looger, L.L. | | Deposit date: | 2011-06-14 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Optimization of a GCaMP calcium indicator for neural activity imaging.
J.Neurosci., 32, 2012
|
|
3SM4
 
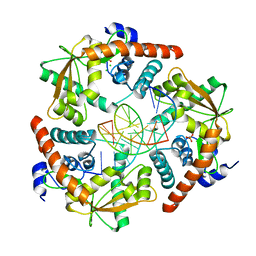 | |
3SG3
 
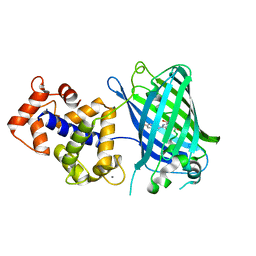 | | Crystal Structure of GCaMP3-D380Y | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Schreiter, E.R, Akerboom, J, Looger, L.L. | | Deposit date: | 2011-06-14 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Optimization of a GCaMP calcium indicator for neural activity imaging.
J.Neurosci., 32, 2012
|
|
3SF8
 
 | |
3SG4
 
 | | Crystal Structure of GCaMP3-D380Y, LP(linker 2) | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Schreiter, E.R, Akerboom, J, Looger, L.L. | | Deposit date: | 2011-06-14 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Optimization of a GCaMP calcium indicator for neural activity imaging.
J.Neurosci., 32, 2012
|
|
3SLP
 
 | |
3VNY
 
 | | Crystal structure of beta-glucuronidase from Acidobacterium capsulatum | | Descriptor: | GLYCEROL, PHOSPHATE ION, beta-GLUCURONIDASE | | Authors: | Momma, M, Fujimoto, Z, Michikawa, M, Ichinose, H, Yoshida, M, Kotake, Y, Biely, P, Tsumuraya, Y, Kaneko, S. | | Deposit date: | 2012-01-18 | | Release date: | 2012-02-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and biochemical characterization of glycoside hydrolase family 79 beta-glucuronidase from Acidobacterium capsulatum
J.Biol.Chem., 287, 2012
|
|
3VJK
 
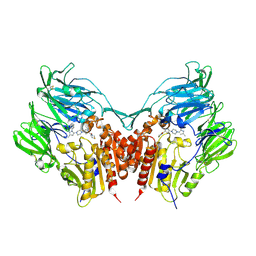 | | Crystal structure of human depiptidyl peptidase IV (DPP-4) in complex with MP-513 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Akahoshi, F, Kishida, H, Miyaguchi, I, Yoshida, T, Ishii, S. | | Deposit date: | 2011-10-24 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Discovery and preclinical profile of teneligliptin (3-[(2S,4S)-4-[4-(3-methyl-1-phenyl-1H-pyrazol-5-yl)piperazin-1-yl]pyrrolidin-2-ylcarbonyl]thiazolidine): A highly potent, selective, long-lasting and orally active dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes
Bioorg.Med.Chem., 20, 2012
|
|
3VKF
 
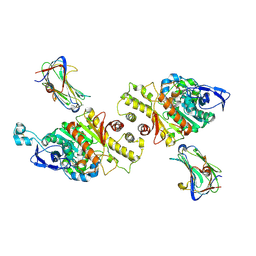 | | Crystal Structure of Neurexin 1beta/Neuroligin 1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neurexin-1-beta, ... | | Authors: | Tanaka, H, Miyazaki, N, Nogi, T, Iwasaki, K, Takagi, J. | | Deposit date: | 2011-11-15 | | Release date: | 2012-08-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Higher-order architecture of cell adhesion mediated by polymorphic synaptic adhesion molecules neurexin and neuroligin.
Cell Rep, 2, 2012
|
|
3US9
 
 | | Crystal Structure of the NCX1 Intracellular Tandem Calcium Binding Domains(CBD12) | | Descriptor: | CALCIUM ION, Sodium/calcium exchanger 1 | | Authors: | Giladi, M, Sasson, Y, Hirsch, J.A, Khananshvili, D. | | Deposit date: | 2011-11-23 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | A common Ca2+-driven interdomain module governs eukaryotic NCX regulation.
Plos One, 7, 2012
|
|
3V6B
 
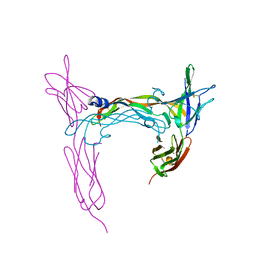 | | VEGFR-2/VEGF-E complex structure | | Descriptor: | VEGF-E, Vascular endothelial growth factor receptor 2 | | Authors: | Brozzo, M.S, Leppanen, V.-M, Winkler, F.K, Kisko, K, Ballmer-Hofer, K. | | Deposit date: | 2011-12-19 | | Release date: | 2012-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Thermodynamic and structural description of allosterically regulated VEGFR-2 dimerization.
Blood, 119, 2012
|
|
3T2D
 
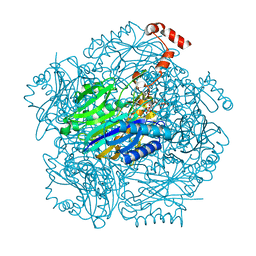 | | Fructose-1,6-bisphosphate aldolase/phosphatase from Thermoproteus neutrophilus, FBP-bound form | | Descriptor: | 1,6-di-O-phosphono-D-fructose, Fructose-1,6-bisphosphate aldolase/phosphatase, MAGNESIUM ION | | Authors: | Du, J, Say, R, Lue, W, Fuchs, G, Einsle, O. | | Deposit date: | 2011-07-22 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Active-site remodelling in the bifunctional fructose-1,6-bisphosphate aldolase/phosphatase.
Nature, 478, 2011
|
|
3THW
 
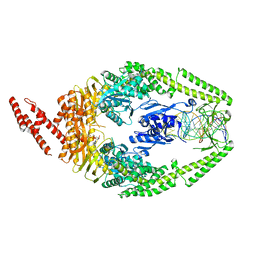 | | Human MutSbeta complexed with an IDL of 4 bases (Loop4) and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA Loop4 hairpin, DNA mismatch repair protein Msh2, ... | | Authors: | Yang, W. | | Deposit date: | 2011-08-19 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Mechanism of mismatch recognition revealed by human MutSbeta bound to unpaired DNA loops
Nat.Struct.Mol.Biol., 19, 2012
|
|
3T0E
 
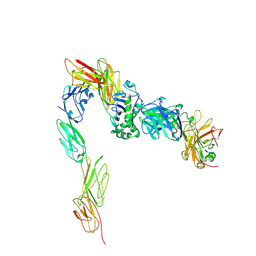 | | Crystal structure of a complete ternary complex of T cell receptor, peptide-MHC and CD4 | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, DRB1-4 beta chain, ... | | Authors: | Yin, Y, Mariuzza, R.A. | | Deposit date: | 2011-07-20 | | Release date: | 2012-03-07 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal structure of a complete ternary complex of T-cell receptor, peptide-MHC, and CD4.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VKH
 
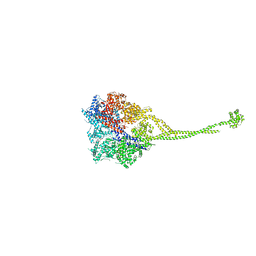 | | X-ray structure of a functional full-length dynein motor domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dynein heavy chain, cytoplasmic | | Authors: | Kon, T, Oyama, T, Shimo-Kon, R, Suto, K, Kurisu, G. | | Deposit date: | 2011-11-16 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The 2.8 A crystal structure of the dynein motor domain
Nature, 484, 2012
|
|
3VO0
 
 | | Crystal structure of beta-glucuronidase from Acidobacterium capsulatum covalent-bonded with 2-deoxy-2-fluoro-D-glucuronic acid | | Descriptor: | 2,4-DINITROPHENOL, 2-deoxy-2-fluoro-alpha-D-glucopyranuronic acid, 2-deoxy-2-fluoro-beta-D-glucopyranuronic acid, ... | | Authors: | Momma, M, Fujimoto, Z, Michikawa, M, Ichinose, H, Jongkees, S, Yoshida, M, Kotake, Y, Biely, P, Tsumuraya, Y, Withers, S, Kaneko, S. | | Deposit date: | 2012-01-18 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biochemical characterization of glycoside hydrolase family 79 beta-glucuronidase from Acidobacterium capsulatum
J.Biol.Chem., 287, 2012
|
|
