4KRG
 
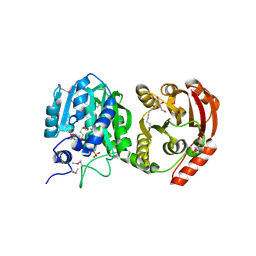 | |
2IAG
 
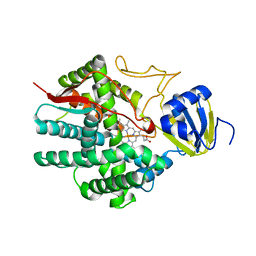 | | Crystal structure of human prostacyclin synthase | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Prostacyclin synthase, SODIUM ION | | Authors: | Chiang, C.-W, Yeh, H.-C, Wang, L.-H, Chan, N.-L. | | Deposit date: | 2006-09-08 | | Release date: | 2006-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Human Prostacyclin Synthase
J.Mol.Biol., 364, 2006
|
|
1JZN
 
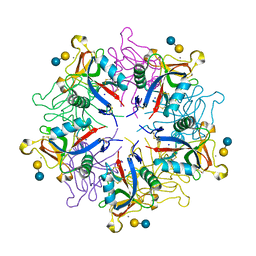 | | crystal structure of a galactose-specific C-type lectin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Galactose-specific lectin, ... | | Authors: | Walker, J.R, Nagar, B, Young, N.M, Hirama, T, Rini, J.M. | | Deposit date: | 2001-09-16 | | Release date: | 2003-07-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray Crystal Structure of a Galactose-Specific C-Type Lectin Possessing a Novel Decameric Quaternary Structure.
Biochemistry, 43, 2004
|
|
4K19
 
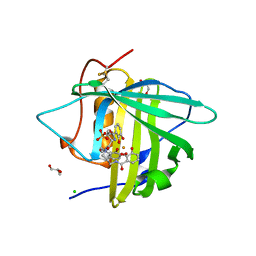 | | The structure of Human Siderocalin bound to the bacterial siderophore fluvibactin | | Descriptor: | (4S,5R)-N,N-bis{3-[(2,3-dihydroxybenzoyl)amino]propyl}-2-(2,3-dihydroxyphenyl)-5-methyl-4,5-dihydro-1,3-oxazole-4-carboxamide, CHLORIDE ION, FE (III) ION, ... | | Authors: | Correnti, C, Clifton, M.C, Strong, R.K. | | Deposit date: | 2013-04-04 | | Release date: | 2013-07-31 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Siderocalin Outwits the Coordination Chemistry of Vibriobactin, a Siderophore of Vibrio cholerae.
Acs Chem.Biol., 8, 2013
|
|
1LCC
 
 | | STRUCTURE OF THE COMPLEX OF LAC REPRESSOR HEADPIECE AND AN 11 BASE-PAIR HALF-OPERATOR DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | DNA (5'-D(*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*T)-3'), Lac Repressor, ... | | Authors: | Chuprina, V.P, Rullmann, J.A.C, Lamerichs, R.M.J.N, Van Boom, J.H, Boelens, R, Kaptein, R. | | Deposit date: | 1993-03-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the complex of lac repressor headpiece and an 11 base-pair half-operator determined by nuclear magnetic resonance spectroscopy and restrained molecular dynamics.
J.Mol.Biol., 234, 1993
|
|
4P0C
 
 | | Crystal Structure of NHERF2 PDZ1 Domain in Complex with LPA2 | | Descriptor: | CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF2/Lysophosphatidic acid receptor 2 chimeric protein, THIOCYANATE ION | | Authors: | Holcomb, J, Jiang, Y, Lu, G, Trescott, L, Brunzelle, J, Sirinupong, N, Li, C, Naren, A, Yang, Z. | | Deposit date: | 2014-02-20 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.339 Å) | | Cite: | Structural insights into PDZ-mediated interaction of NHERF2 and LPA2, a cellular event implicated in CFTR channel regulation.
Biochem.Biophys.Res.Commun., 446, 2014
|
|
1M0P
 
 | | Structure of Dialkylglycine Decarboxylase Complexed with 1-Amino-1-phenylethanephosphonate | | Descriptor: | (1R)-1-[((1E)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYLENE)AMINO]-1-PHENYLETHYLPHOSPHONIC ACID, 2,2-Dialkylglycine Decarboxylase, POTASSIUM ION, ... | | Authors: | Liu, W, Rogers, C.J, Fisher, A.J, Toney, M.D. | | Deposit date: | 2002-06-13 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Aminophosphonate Inhibitors of Dialkylglycine Decarboxylase: Structural Basis for Slow Binding Inhibition
Biochemistry, 41, 2002
|
|
1M0Q
 
 | | Structure of Dialkylglycine Decarboxylase Complexed with S-1-aminoethanephosphonate | | Descriptor: | (1S)-1-[((1E)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYLENE)AMINO]ETHYLPHOSPHONIC ACID, 2,2-Dialkylglycine Decarboxylase, POTASSIUM ION, ... | | Authors: | Liu, W, Rogers, C.J, Fisher, A.J, Toney, M.D. | | Deposit date: | 2002-06-13 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aminophosphonate Inhibitors of Dialkylglycine Decarboxylase: Structural Basis for Slow Binding Inhibition
Biochemistry, 41, 2002
|
|
2GJP
 
 | | Structure of Bacillus halmapalus alpha-amylase, crystallized with the substrate analogue acarbose and maltose | | Descriptor: | 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lyhne-Iversen, L, Hobley, T.J, Kaasgaard, S.G, Harris, P. | | Deposit date: | 2006-03-31 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Bacillus halmapalus alpha-amylase crystallized with and without the substrate analogue acarbose and maltose.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
4OFI
 
 | | Crystal Structure of Duf (Kirre) D1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Kin of irre, isoform A, ... | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
1IP6
 
 | | G127A HUMAN LYSOZYME | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2001-04-20 | | Release date: | 2001-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of amino acid residues in left-handed helical conformation for the conformational stability of a protein.
Proteins, 45, 2001
|
|
4MVQ
 
 | | Structural Basis for Ca2+ Selectivity of a Voltage-gated Calcium Channel | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein | | Authors: | Tang, L, Gamal El-Din, T.M, Payandeh, J, Gilbert, Q.M, Heard, T.M, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2013-09-24 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for Ca2+ selectivity of a voltage-gated calcium channel.
Nature, 505, 2014
|
|
1LRA
 
 | | CRYSTALLOGRAPHIC STUDY OF GLU 58 ALA RNASE T1(ASTERISK)2'-GUANOSINE MONOPHOSPHATE AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1, SODIUM ION | | Authors: | Pletinckx, J, Steyaert, J, Choe, H.-W, Heinemann, U, Wyns, L. | | Deposit date: | 1993-10-01 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic study of Glu58Ala RNase T1 x 2'-guanosine monophosphate at 1.9-A resolution.
Biochemistry, 33, 1994
|
|
1LCD
 
 | | STRUCTURE OF THE COMPLEX OF LAC REPRESSOR HEADPIECE AND AN 11 BASE-PAIR HALF-OPERATOR DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | DNA (5'-D(*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*T)-3'), Lac Repressor, ... | | Authors: | Chuprina, V.P, Rullmann, J.A.C, Lamerichs, R.M.J.N, Van Boom, J.H, Boelens, R, Kaptein, R. | | Deposit date: | 1993-03-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the complex of lac repressor headpiece and an 11 base-pair half-operator determined by nuclear magnetic resonance spectroscopy and restrained molecular dynamics.
J.Mol.Biol., 234, 1993
|
|
4IRZ
 
 | | Crystal structure of A4b7 headpiece complexed with Fab Natalizumab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yu, Y, Schurpf, T, Springer, T.A. | | Deposit date: | 2013-01-15 | | Release date: | 2013-09-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | How natalizumab binds and antagonizes alpha 4 integrins.
J.Biol.Chem., 288, 2013
|
|
2HYA
 
 | | HYALURONIC ACID, MOLECULAR CONFORMATIONS AND INTERACTIONS IN TWO SODIUM SALTS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranuronic acid, SODIUM ION | | Authors: | Arnott, S. | | Deposit date: | 1977-11-20 | | Release date: | 1980-03-28 | | Last modified: | 2024-02-21 | | Method: | FIBER DIFFRACTION (3 Å) | | Cite: | Hyaluronic acid: molecular conformations and interactions in two sodium salts.
J.Mol.Biol., 95, 1975
|
|
4IWV
 
 | | Crystals structure of Human Glucokinase in complex with small molecule activator | | Descriptor: | (2S)-2-{[1-(2-chlorophenyl)-1H-pyrazolo[3,4-d]pyrimidin-4-yl]oxy}-N-(5-chloropyridin-2-yl)-3-(2-hydroxyethoxy)propanamide, Glucokinase isoform 3, SODIUM ION, ... | | Authors: | Ogg, D.J, Hargreaves, D, Gerhardt, S, Flavell, L, McAlister, M. | | Deposit date: | 2013-01-24 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Optimising pharmacokinetics of glucokinase activators with matched triplicate design sets the discovery of AZD3651 and AZD9485
To be Published
|
|
4HMK
 
 | | Crystal structure of LeuT-E290S with bound Br | | Descriptor: | BROMIDE ION, LEUCINE, SODIUM ION, ... | | Authors: | Kantcheva, A.K, Quick, M, Shi, L, Winther, A.M.L, Stolzenberg, S, Weinstein, H, Javitch, J.A, Nissen, P. | | Deposit date: | 2012-10-18 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The chloride binding site of Neurotransmitter Sodium Symporters
Proc.Natl.Acad.Sci.USA, 2013
|
|
4I9X
 
 | |
1LZ8
 
 | | LYSOZYME PHASED ON ANOMALOUS SIGNAL OF SULFURS AND CHLORINES | | Descriptor: | CHLORIDE ION, PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Dauter, Z, Dauter, M, De La Fortelle, E, Bricogne, G, Sheldrick, G.M. | | Deposit date: | 1999-03-14 | | Release date: | 1999-05-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Can anomalous signal of sulfur become a tool for solving protein crystal structures?
J.Mol.Biol., 289, 1999
|
|
4I7Y
 
 | | Crystal Structure of Human Alpha Thrombin in Complex with a 27-mer Aptamer Bound to Exosite II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, DNA (27-MER), ... | | Authors: | Pica, A, Russo Krauss, I, Merlino, A, Mazzarella, L, Sica, F. | | Deposit date: | 2012-12-01 | | Release date: | 2013-10-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Duplex-quadruplex motifs in a peculiar structural organization cooperatively contribute to thrombin binding of a DNA aptamer.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4P30
 
 | | Structure of NavMS mutant in presence of PI1 compound | | Descriptor: | DODECAETHYLENE GLYCOL, HEGA-10, Ion transport protein, ... | | Authors: | Bagneris, C, Naylor, C.E, Wallace, B.A. | | Deposit date: | 2014-03-05 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Prokaryotic NavMs channel as a structural and functional model for eukaryotic sodium channel antagonism.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4PA6
 
 | | Structure of NavMS pore and C-terminal domain crystallised in the presence of channel blocking compound | | Descriptor: | DODECAETHYLENE GLYCOL, HEGA-10, Ion transport protein, ... | | Authors: | Naylor, C.E, Bagneris, C, Wallace, B.A. | | Deposit date: | 2014-04-07 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Prokaryotic NavMs channel as a structural and functional model for eukaryotic sodium channel antagonism.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4HOD
 
 | | Crystal structure of LeuT-E290S with bound Cl | | Descriptor: | CHLORIDE ION, LEUCINE, SODIUM ION, ... | | Authors: | Kantcheva, A.K, Quick, M, Shi, L, Winther, A.M.L, Stolzenberg, S, Weinstein, H, Javitch, J.A, Nissen, P. | | Deposit date: | 2012-10-22 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The chloride binding site of Neurotransmitter Sodium Symporters
Proc.Natl.Acad.Sci.USA, 2013
|
|
1IP4
 
 | | G72A HUMAN LYSOZYME | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2001-04-20 | | Release date: | 2001-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of amino acid residues in left-handed helical conformation for the conformational stability of a protein.
Proteins, 45, 2001
|
|
