9FAL
 
 | | Gcase in complex with small molecule inhibitor 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tisi, D, Cleasby, A. | | Deposit date: | 2024-05-10 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Fragment-Based Discovery of a Series of Allosteric-Binding Site Modulators of beta-Glucocerebrosidase.
J.Med.Chem., 67, 2024
|
|
7WH9
 
 | | holo structure of emodin 1-OH O-methyltransferase complex with emodin and S-Adenosyl-L-homocysteine | | Descriptor: | 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, O-methyltransferase gedA, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Liang, Y.J, Lu, X.F, Qi, F.F, Xue, Y.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Characterization and Structural Analysis of Emodin- O -Methyltransferase from Aspergillus terreus.
J.Agric.Food Chem., 70, 2022
|
|
4MSE
 
 | | Crystal structure of PDE10A2 with fragment ZT1597 (2-({[(2S)-2-methyl-2,3-dihydro-1,3-benzothiazol-5-yl]oxy}methyl)quinoline) | | Descriptor: | 2-{[(2-methyl-1,3-benzothiazol-5-yl)oxy]methyl}quinoline, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
4MWO
 
 | | Crystal structure of human mitochondrial 5'(3')-deoxyribonucleotidase in complex with the inhibitor CPB-T | | Descriptor: | 1-{3,5-O-[(4-carboxyphenyl)(phosphono)methylidene]-2-deoxy-beta-D-threo-pentofuranosyl}-5-methylpyrimidine-2,4(1H,3H)-dione, 5'(3')-deoxyribonucleotidase, mitochondrial, ... | | Authors: | Pachl, P, Rezacova, P, Brynda, J. | | Deposit date: | 2013-09-25 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Conformationally constrained nucleoside phosphonic acids - potent inhibitors of human mitochondrial and cytosolic 5'(3')-nucleotidases.
Org.Biomol.Chem., 12, 2014
|
|
7TNC
 
 | | M13F/G116F Pseudomonas aeruginosa azurin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Azurin, CHLORIDE ION, ... | | Authors: | Liu, Y, Lu, Y. | | Deposit date: | 2022-01-20 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Basis for the Effects of Phenylalanine on Tuning the Reduction Potential of Type 1 Copper in Azurin.
Inorg.Chem., 62, 2023
|
|
9F9Y
 
 | | SARS-CoV-2 BA-2.87.1 Spike ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Ren, J, Stuart, D.I, Duyvesteyn, H.M.E. | | Deposit date: | 2024-05-09 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Concerted deletions eliminate a neutralizing supersite in SARS-CoV-2 BA.2.87.1 spike.
Structure, 32, 2024
|
|
5JPA
 
 | | Hexameric HIV-1 CA H12Y mutant | | Descriptor: | Capsid Protein P24 | | Authors: | Jacques, D.A, James, L.C. | | Deposit date: | 2016-05-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | HIV-1 uses dynamic capsid pores to import nucleotides and fuel encapsidated DNA synthesis.
Nature, 536, 2016
|
|
6EPI
 
 | |
8JYP
 
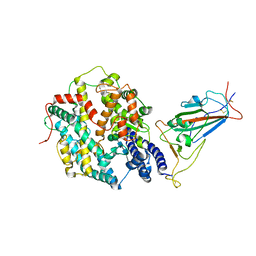 | | Structure of SARS-CoV-2 XBB.1.5 spike RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron XBB.1.5 variant.
Nat Commun, 15, 2024
|
|
8JYN
 
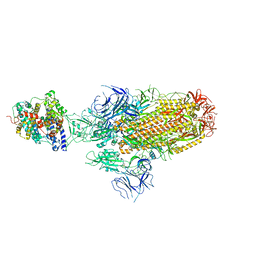 | | Structure of SARS-CoV-2 XBB.1.5 spike glycoprotein in complex with ACE2 (1-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron XBB.1.5 variant.
Nat Commun, 15, 2024
|
|
8JYO
 
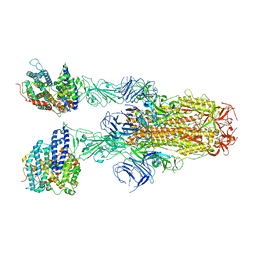 | | Structure of SARS-CoV-2 XBB.1.5 spike glycoprotein in complex with ACE2 (2-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron XBB.1.5 variant.
Nat Commun, 15, 2024
|
|
9BLY
 
 | |
7WI1
 
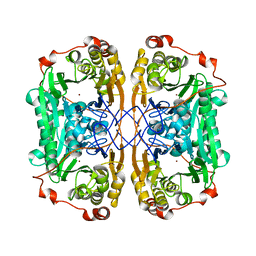 | |
5HGM
 
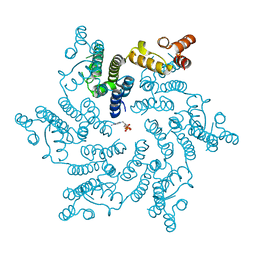 | | Hexameric HIV-1 CA in complex with dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Capsid protein P24 | | Authors: | Jacques, D.A, James, L.C. | | Deposit date: | 2016-01-08 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | HIV-1 uses dynamic capsid pores to import nucleotides and fuel encapsidated DNA synthesis.
Nature, 536, 2016
|
|
6A1H
 
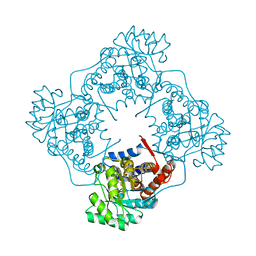 | | Mandelate oxidase mutant-Y128F with 5-deazariboflavin mononucleotide | | Descriptor: | 1-deoxy-1-(7,8-dimethyl-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-5-O-phosphono-D-ribitol, 4-hydroxymandelate oxidase | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-07 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The flavin mononucleotide cofactor in alpha-hydroxyacid oxidases exerts its electrophilic/nucleophilic duality in control of the substrate-oxidation level.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5HSV
 
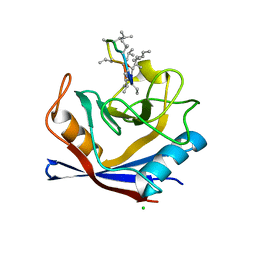 | | X-Ray structure of a CypA-Alisporivir complex at 1.5 angstrom resolution | | Descriptor: | Alisporivir, CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Dujardin, M, Bouckaert, J, Rucktooa, P, Hanoulle, X. | | Deposit date: | 2016-01-26 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray structure of alisporivir in complex with cyclophilin A at 1.5 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5JY5
 
 | | Crystal structure of Thioredoxin 1 from Cryptococcus neoformans at 1.8 Angstroms resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Bravo-Chaucanes, C.P, Abadio, A.K.R, Kioshima, E.S, Felipe, M.S.S, Barbosa, J.A.R.G. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Thioredoxin 1 from Cryptococcus neoformans at 1.8 Angstroms resolution
To Be Published
|
|
6A1E
 
 | | Crystal structure of a synthase 1 from Santalum album in complex with ligand fps | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CARBONATE ION, GLYCEROL, ... | | Authors: | Han, X, Ko, T.P, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-06-07 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of a synthase 1 from Santalum album in complex with ligand
To Be Published
|
|
6A1R
 
 | | Mandelate oxidase mutant-Y128F with the N5-phenylacetyl-FMN adduct | | Descriptor: | 1-deoxy-1-[7,8-dimethyl-2,4-dioxo-5-(phenylacetyl)-1,3,4,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]-5-O-phosphono-D-ribitol, 4-hydroxymandelate oxidase, MAGNESIUM ION | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-08 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | The flavin mononucleotide cofactor in alpha-hydroxyacid oxidases exerts its electrophilic/nucleophilic duality in control of the substrate-oxidation level.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5HA9
 
 | | Crystal structure-based design and disovery of a novel PARP1 antiagonist (BL-PA10) that induces apoptosis and inhibits metastasis in triple negative breast cancer | | Descriptor: | Amitriptyline, GLYCEROL, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Fu, L, Peng, H, Zhang, L, Ouyang, L. | | Deposit date: | 2015-12-30 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (4.01 Å) | | Cite: | Crystal structure-based discovery of a novel synthesized PARP1 inhibitor (OL-1) with apoptosis-inducing mechanisms in triple-negative breast cancer.
Sci Rep, 6, 2016
|
|
6TKN
 
 | | Tankyrase 2 in complex with an inhibitor (OM-1000) | | Descriptor: | Tankyrase-2, ZINC ION, ~{N}-[3-[4-(2-chlorophenyl)-5-pyrimidin-4-yl-1,2,4-triazol-3-yl]-1-bicyclo[1.1.1]pentanyl]pyridine-2-carboxamide | | Authors: | Sowa, S.T, Lehtio, L. | | Deposit date: | 2019-11-28 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Preclinical Lead Optimization of a 1,2,4-Triazole Based Tankyrase Inhibitor.
J.Med.Chem., 63, 2020
|
|
5DFS
 
 | | Crystal structure of Spider Monkey Cytochrome C at 1.15 Angstrom | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cytochrome c, ... | | Authors: | MOU, T.C, McClelland, L.J, Jeakins-Cooley, M.E, Goldes, M.E, SPRANG, S.R, BOWLER, B.E. | | Deposit date: | 2015-08-27 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Disruption of a hydrogen bond network in human versus spider monkey cytochrome c affects heme crevice stability.
J.Inorg.Biochem., 158, 2016
|
|
3L3V
 
 | | Structure of HIV-1 integrase core domain in complex with sucrose | | Descriptor: | CADMIUM ION, POL polyprotein, SULFATE ION, ... | | Authors: | Wielens, J, Chalmers, D.K, Scanlon, M.J, Parker, M.W. | | Deposit date: | 2009-12-18 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the HIV-1 integrase core domain in complex with sucrose reveals details of an allosteric inhibitory binding site
Febs Lett., 584, 2010
|
|
3ZJ7
 
 | | Crystal structure of strictosidine glucosidase in complex with inhibitor-1 | | Descriptor: | (1R,2S,3S,4R,5R)-4-(cyclohexylamino)-5-(hydroxymethyl)cyclopentane-1,2,3-triol, STRICTOSIDINE-O-BETA-D-GLUCOSIDASE | | Authors: | Xia, L, Lin, H, Panjikar, S, Ruppert, M, Castiglia, A, Rajendran, C, Wang, M, Schuebel, H, Warzecha, H, Jaeger, V, Stoeckigt, J. | | Deposit date: | 2013-01-17 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand Structures of Synthetic Deoxa-Pyranosylamines with Raucaffricine and Strictosidine Glucosidases Provide Structural Insights Into Their Binding and Inhibitory Behaviours.
J.Enzyme.Inhib.Med.Chem., 30, 2015
|
|
3ZM9
 
 | | The mechanism of allosteric coupling in choline kinase a1 revealed by a rationally designed inhibitor | | Descriptor: | 1-(4-(4-(4-((6-amino-9H-purin-9-yl)methyl)phenyl)butyl)benzyl)-4- (dimethylamino)pyridinium, CHOLINE KINASE ALPHA | | Authors: | Sahun-Roncero, M, Rubio-Ruiz, B, Saladino, G, Conejo-Garcia, A, Espinosa, A, Velazquez-Campoy, A, Gervasio, F.L, Entrena, A, Hurtado-Guerrero, R. | | Deposit date: | 2013-02-06 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Mechanism of Allosteric Coupling in Choline Kinase A1 Revealed by a Rationally Designed Inhibitor
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
