5ZHM
 
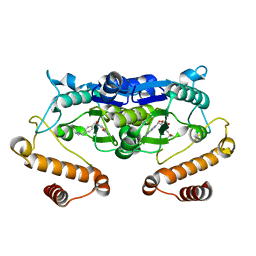 | | Crystal structure of TrmD from Pseudomonas aeruginosa in complex with active-site inhibitor | | Descriptor: | N-({4-[(diethylamino)methyl]phenyl}methyl)-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidine-5-carboxamide, S-ADENOSYLMETHIONINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Zhong, W, Pasunooti, K.K, Balamkundu, S, Wong, Y.W, Nah, Q, Liu, C.F, Lescar, J, Dedon, P.C. | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Thienopyrimidinone Derivatives That Inhibit Bacterial tRNA (Guanine37-N1)-Methyltransferase (TrmD) by Restructuring the Active Site with a Tyrosine-Flipping Mechanism.
J.Med.Chem., 62, 2019
|
|
7XOE
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Prefusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
5ZHK
 
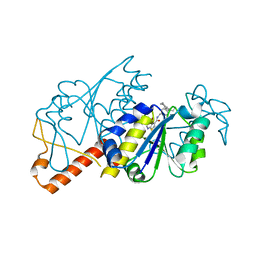 | | Crystal structure of TrmD from Mycobacterium tuberculosis in complex with active-site inhibitor | | Descriptor: | N-[(4-{[cyclohexyl(ethyl)amino]methyl}phenyl)methyl]-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidine-5-carboxamide, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Zhong, W, Pasunooti, K.K, Balamkundu, S, Wong, Y.W, Nah, Q, Liu, C.F, Lescar, J, Dedon, P.C. | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Thienopyrimidinone Derivatives That Inhibit Bacterial tRNA (Guanine37-N1)-Methyltransferase (TrmD) by Restructuring the Active Site with a Tyrosine-Flipping Mechanism.
J.Med.Chem., 62, 2019
|
|
5ZHN
 
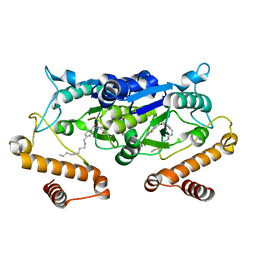 | | Crystal structure of TrmD from Pseudomonas aeruginosa in complex with active-site inhibitor | | Descriptor: | N-({4-[(octylamino)methyl]phenyl}methyl)-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidine-5-carboxamide, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Zhong, W, Pasunooti, K.K, Balamkundu, S, Wong, Y.W, Nah, Q, Liu, C.F, Lescar, J, Dedon, P.C. | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Thienopyrimidinone Derivatives That Inhibit Bacterial tRNA (Guanine37-N1)-Methyltransferase (TrmD) by Restructuring the Active Site with a Tyrosine-Flipping Mechanism.
J.Med.Chem., 62, 2019
|
|
5ZHL
 
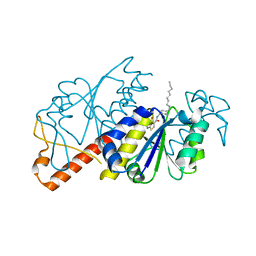 | | Crystal structure of TrmD from Mycobacterium tuberculosis in complex with active-site inhibitor | | Descriptor: | N-({4-[(octylamino)methyl]phenyl}methyl)-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidine-5-carboxamide, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Zhong, W, Pasunooti, K.K, Balamkundu, S, Wong, Y.W, Nah, Q, Liu, C.F, Lescar, J, Dedon, P.C. | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Thienopyrimidinone Derivatives That Inhibit Bacterial tRNA (Guanine37-N1)-Methyltransferase (TrmD) by Restructuring the Active Site with a Tyrosine-Flipping Mechanism.
J.Med.Chem., 62, 2019
|
|
5ZHJ
 
 | | Crystal structure of TrmD from Mycobacterium tuberculosis in complex with S-adenosyl homocysteine (SAH) | | Descriptor: | GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Zhong, W, Pasunooti, K.K, Balamkundu, S, Wong, Y.W, Nah, Q, Liu, C.F, Lescar, J, Dedon, P.C. | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Thienopyrimidinone Derivatives That Inhibit Bacterial tRNA (Guanine37-N1)-Methyltransferase (TrmD) by Restructuring the Active Site with a Tyrosine-Flipping Mechanism.
J.Med.Chem., 62, 2019
|
|
1LBA
 
 | |
2YX1
 
 | | Crystal structure of M.jannaschii tRNA m1G37 methyltransferase | | Descriptor: | Hypothetical protein MJ0883, SINEFUNGIN, ZINC ION | | Authors: | Goto-Ito, S, Ito, T, Ishii, R, Bessho, Y, Yokoyama, S. | | Deposit date: | 2007-04-23 | | Release date: | 2008-04-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of archaeal tRNA(m(1)G37)methyltransferase aTrm5.
Proteins, 72, 2008
|
|
3HGA
 
 | |
2MFD
 
 | |
3O4N
 
 | |
3O4Q
 
 | |
2ITB
 
 | |
1S0U
 
 | | eIF2gamma apo | | Descriptor: | Translation initiation factor 2 gamma subunit, ZINC ION | | Authors: | Roll-Mecak, A, Alone, P, Cao, C, Dever, T.E, Burley, S.K. | | Deposit date: | 2004-01-04 | | Release date: | 2004-01-20 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray Structure of Translation Initiation Factor eIF2gamma: IMPLICATIONS FOR tRNA AND eIF2alpha BINDING.
J.Biol.Chem., 279, 2004
|
|
1KU7
 
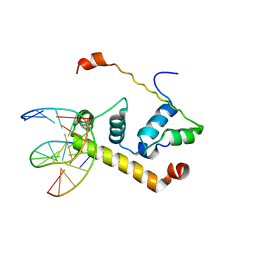 | | Crystal Structure of Thermus aquatics RNA Polymerase SigmaA Subunit Region 4 Bound to-35 Element DNA | | Descriptor: | 5'-D(*CP*CP*TP*TP*GP*AP*CP*AP*AP*AP*G)-3', 5'-D(*CP*CP*TP*TP*TP*GP*TP*CP*AP*AP*G)-3', sigma factor sigA | | Authors: | Campbell, E.A, Muzzin, O, Chlenov, M, Sun, J.L, Olson, C.A, Weinman, O, Trester-Zedlitz, M.L, Darst, S.A. | | Deposit date: | 2002-01-21 | | Release date: | 2002-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the bacterial RNA polymerase promoter specificity sigma subunit.
Mol.Cell, 9, 2002
|
|
1KU3
 
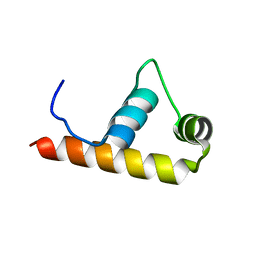 | | Crystal Structure of Thermus aquaticus RNA Polymerase Sigma Subunit Fragment, Region 4 | | Descriptor: | sigma factor sigA | | Authors: | Campbell, E.A, Muzzin, O, Chlenov, M, Sun, J.L, Olson, C.A, Weinman, O, Trester-Zedlitz, M.L, Darst, S.A. | | Deposit date: | 2002-01-21 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the bacterial RNA polymerase promoter specificity sigma subunit.
Mol.Cell, 9, 2002
|
|
8OQX
 
 | | Crystal structure of Tannerella forsythia MurNAc kinase MurK with a phosphate analogue | | Descriptor: | 1,2-ETHANEDIOL, ATPase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gogler, K, Fink, P, Stasiak, A.C, Stehle, T, Zocher, G. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | N-acetylmuramic acid recognition by MurK kinase from the MurNAc auxotrophic oral pathogen Tannerella forsythia.
J.Biol.Chem., 299, 2023
|
|
8OW7
 
 | | Crystal structure of Tannerella forsythia sugar kinase K1058 in complex with N-acetylmuramic acid (MurNAc) | | Descriptor: | N-acetyl-beta-muramic acid, N-acetylglucosamine kinase, SULFATE ION | | Authors: | Stasiak, A.C, Gogler, K, Fink, P, Stehle, T, Zocher, G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-08-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | N-acetylmuramic acid recognition by MurK kinase from the MurNAc auxotrophic oral pathogen Tannerella forsythia.
J.Biol.Chem., 299, 2023
|
|
8OQW
 
 | | Crystal structure of Tannerella forsythia MurNAc kinase MurK | | Descriptor: | ATPase, GLYCEROL, SULFATE ION | | Authors: | Gogler, K, Fink, P, Stasiak, A.C, Stehle, T, Zocher, G. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | N-acetylmuramic acid recognition by MurK kinase from the MurNAc auxotrophic oral pathogen Tannerella forsythia.
J.Biol.Chem., 299, 2023
|
|
1KU2
 
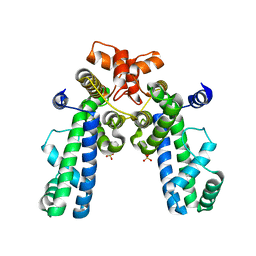 | | Crystal Structure of Thermus aquaticus RNA Polymerase Sigma Subunit Fragment Containing Regions 1.2 to 3.1 | | Descriptor: | SULFATE ION, sigma factor sigA | | Authors: | Campbell, E.A, Muzzin, O, Chlenov, M, Sun, J.L, Olson, C.A, Weinman, O, Trester-Zedlitz, M.L, Darst, S.A. | | Deposit date: | 2002-01-21 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the bacterial RNA polymerase promoter specificity sigma subunit.
Mol.Cell, 9, 2002
|
|
2OXC
 
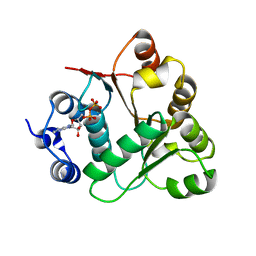 | | Human DEAD-box RNA helicase DDX20, DEAD domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Probable ATP-dependent RNA helicase DDX20 | | Authors: | Karlberg, T, Ogg, D, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Hogbom, M, Johansson, I, Kotenyova, T, Lehtio, L, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-20 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Human DEAD-box RNA helicase DDX20
To be Published
|
|
2X0L
 
 | | Crystal structure of a neuro-specific splicing variant of human histone lysine demethylase LSD1. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, HISTONE H3 PEPTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Zibetti, C, Adamo, A, Binda, C, Forneris, F, Verpelli, C, Ginelli, E, Mattevi, A, Sala, C, Battaglioli, E. | | Deposit date: | 2009-12-15 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Alternative Splicing of the Histone Demethylase Lsd1/Kdm1 Contributes to the Modulation of Neurite Morphogenesis in the Mammalian Nervous System.
J.Neurosci., 30, 2010
|
|
2X67
 
 | | The ternary complex of PrnB (the second enzyme in pyrrolnitrin biosynthesis pathway), tryptophan and cyanide | | Descriptor: | CYANIDE ION, PRNB, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhu, X, van pee, K.-H, Naismith, J.H. | | Deposit date: | 2010-02-15 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The Ternary Complex of Prnb (the Second Enzyme in Pyrrolnitrin Biosynthesis Pathway), Tryptophan and Cyanide Yields New Mechanistic Insights Into the Indolamine Dioxygenase Superfamily.
J.Biol.Chem., 285, 2010
|
|
1XZP
 
 | | Structure of the GTP-binding protein TrmE from Thermotoga maritima | | Descriptor: | Probable tRNA modification GTPase trmE, SULFATE ION | | Authors: | Scrima, A, Vetter, I.R, Armengod, M.E, Wittinghofer, A. | | Deposit date: | 2004-11-12 | | Release date: | 2005-01-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of the TrmE GTP-binding protein and its implications for tRNA modification
Embo J., 24, 2005
|
|
1XZQ
 
 | | Structure of the GTP-binding protein TrmE from Thermotoga maritima complexed with 5-formyl-THF | | Descriptor: | N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, Probable tRNA modification GTPase trmE | | Authors: | Scrima, A, Vetter, I.R, Armengod, M.E, Wittinghofer, A. | | Deposit date: | 2004-11-12 | | Release date: | 2005-01-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of the TrmE GTP-binding protein and its implications for tRNA modification
Embo J., 24, 2005
|
|
