3SML
 
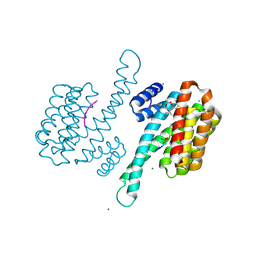 | | Crystal structure of human 14-3-3 sigma C38N/N166H in complex with TASK-3 peptide and stabilizer Fusicoccin A aglycone | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Fusicoccin A aglycone, ... | | Authors: | Anders, C, Schumacher, B, Ottmann, C. | | Deposit date: | 2011-06-28 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A semisynthetic fusicoccane stabilizes a protein-protein interaction and enhances the expression of K+ channels at the cell surface.
Chem. Biol., 20, 2013
|
|
3UX0
 
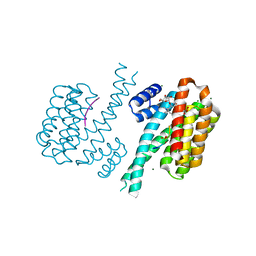 | | Crystal structure of human 14-3-3 sigma in complex with TASK-3 peptide and stabilizer Fusicoccin H | | Descriptor: | (4R,5R,6R,6aS,9S,9aE,10aR)-5-hydroxy-9-(hydroxymethyl)-6,10a-dimethyl-3-(propan-2-yl)-1,2,4,5,6,6a,7,8,9,10a-decahydrodicyclopenta[a,d][8]annulen-4-yl alpha-D-gulopyranoside, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Thiel, P, Bartel, M, Anders, C, Higuchi, Y, Schumacher, B, Kato, N, Ottmann, C. | | Deposit date: | 2011-12-03 | | Release date: | 2013-01-02 | | Last modified: | 2013-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A semisynthetic fusicoccane stabilizes a protein-protein interaction and enhances the expression of k(+) channels at the cell surface.
Chem.Biol., 20, 2013
|
|
1PBD
 
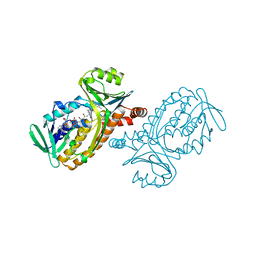 | | CRYSTAL STRUCTURES OF WILD-TYPE P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE, 2,4-DIHYDROXYBENZOATE AND 2-HYDROXY-4-AMINOBENZOATE AND OF THE TRY222ALA MUTANT, COMPLEXED WITH 2-HYDROXY-4-AMINOBENZOATE. EVIDENCE FOR A PROTON CHANNEL AND A NEW BINDING MODE OF THE FLAVIN RING | | Descriptor: | 4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Schreuder, H.A, Mattevi, A, Hol, W.G.J. | | Deposit date: | 1994-07-06 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring
Biochemistry, 33, 1994
|
|
1PBF
 
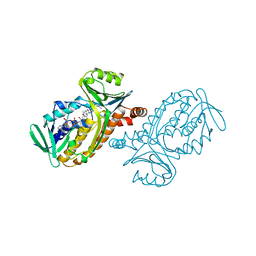 | | CRYSTAL STRUCTURES OF WILD-TYPE P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE, 2,4-DIHYDROXYBENZOATE AND 2-HYDROXY-4-AMINOBENZOATE AND OF THE TRY222ALA MUTANT, COMPLEXED WITH 2-HYDROXY-4-AMINOBENZOATE. EVIDENCE FOR A PROTON CHANNEL AND A NEW BINDING MODE OF THE FLAVIN RING | | Descriptor: | 2-HYDROXY-4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Schreuder, H.A, Van Der Bolt, F.J.T, Van Berkel, W.J.H. | | Deposit date: | 1994-07-06 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring
Biochemistry, 33, 1994
|
|
1PBB
 
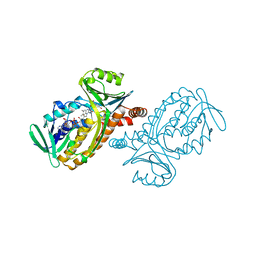 | | CRYSTAL STRUCTURES OF WILD-TYPE P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE, 2,4-DIHYDROXYBENZOATE AND 2-HYDROXY-4-AMINOBENZOATE AND OF THE TRY222ALA MUTANT, COMPLEXED WITH 2-HYDROXY-4-AMINOBENZOATE. EVIDENCE FOR A PROTON CHANNEL AND A NEW BINDING MODE OF THE FLAVIN RING | | Descriptor: | 2,4-DIHYDROXYBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Schreuder, H.A, Mattevi, A, Hol, W.G.J. | | Deposit date: | 1994-07-06 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring
Biochemistry, 33, 1994
|
|
1PBC
 
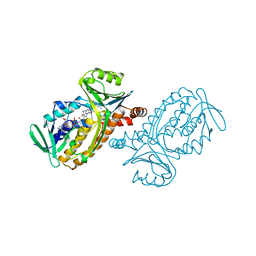 | | CRYSTAL STRUCTURES OF WILD-TYPE P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE, 2,4-DIHYDROXYBENZOATE AND 2-HYDROXY-4-AMINOBENZOATE AND OF THE TRY222ALA MUTANT, COMPLEXED WITH 2-HYDROXY-4-AMINOBENZOATE. EVIDENCE FOR A PROTON CHANNEL AND A NEW BINDING MODE OF THE FLAVIN RING | | Descriptor: | 2-HYDROXY-4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Schreuder, H.A, Van Der Bolt, F.J.T, Van Berkel, W.J.H. | | Deposit date: | 1994-07-06 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring
Biochemistry, 33, 1994
|
|
8CUA
 
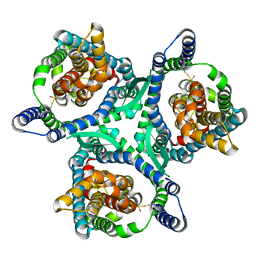 | |
3V7T
 
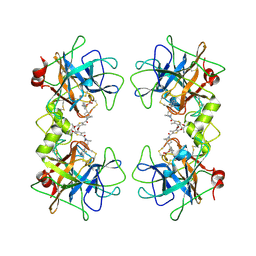 | |
7ZRL
 
 | | Cryo-EM map of the unphosphorylated KdpFABC complex in the E2-P conformation, under turnover conditions | | Descriptor: | POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, Potassium-transporting ATPase KdpC subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
2M0Q
 
 | | Solution NMR analysis of intact KCNE2 in detergent micelles demonstrate a straight transmembrane helix | | Descriptor: | Potassium voltage-gated channel subfamily E member 2 | | Authors: | Lai, C, Li, P, Chen, L, Zhang, L, Wu, F, Tian, C. | | Deposit date: | 2012-11-01 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Differential modulations of KCNQ1 by auxiliary proteins KCNE1 and KCNE2.
Sci Rep, 4, 2014
|
|
2CK5
 
 | |
3EXG
 
 | | Crystal structure of the pyruvate dehydrogenase (E1p) component of human pyruvate dehydrogenase complex | | Descriptor: | POTASSIUM ION, Pyruvate dehydrogenase E1 component subunit alpha, somatic form, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Machius, M, Li, J, Chuang, D.T. | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops.
Structure, 16, 2008
|
|
2CK4
 
 | |
6V4J
 
 | | Structure of TrkH-TrkA in complex with ATP | | Descriptor: | Potassium uptake protein TrkA, Trk system potassium uptake protein TrkH | | Authors: | Zhou, M, Zhang, H. | | Deposit date: | 2019-11-27 | | Release date: | 2020-02-12 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | TrkA undergoes a tetramer-to-dimer conversion to open TrkH which enables changes in membrane potential.
Nat Commun, 11, 2020
|
|
4WWS
 
 | |
2RTZ
 
 | | Solution structure of MarkTX-7 | | Descriptor: | Potassium channel blocker MarkTX-7 | | Authors: | Umetsu, Y, Ohki, S. | | Deposit date: | 2013-10-17 | | Release date: | 2014-10-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of MarkTX-7
To be Published
|
|
2MLD
 
 | |
1WJX
 
 | | Crystal sturucture of TT0801 from Thermus thermophilus | | Descriptor: | POTASSIUM ION, SsrA-binding protein | | Authors: | Bessho, Y, Shibata, R, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-29 | | Release date: | 2004-11-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for functional mimicry of long-variable-arm tRNA by transfer-messenger RNA.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6AY8
 
 | |
6ATM
 
 | |
2MLA
 
 | |
6GSH
 
 | | Feline Calicivirus Strain F9 | | Descriptor: | POTASSIUM ION, VP1 | | Authors: | Conley, M.J, Bhella, D. | | Deposit date: | 2018-06-14 | | Release date: | 2019-01-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Calicivirus VP2 forms a portal-like assembly following receptor engagement.
Nature, 565, 2019
|
|
5UM4
 
 | |
6CI1
 
 | |
3ETQ
 
 | |
