7Z0F
 
 | | CPAP:S-TUBULIN:IIH5 ALPHAREP COMPLEX | | Descriptor: | Centromere protein J, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gigant, B, Campanacci, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Structural convergence for tubulin binding of CPAP and vinca domain microtubule inhibitors.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6EWY
 
 | | RipA Peptidoglycan hydrolase (Rv1477, Mycobacterium tuberculosis) N-terminal domain | | Descriptor: | Peptidoglycan endopeptidase RipA | | Authors: | Schnell, R, Steiner, E.M, Schneider, G, Guy, J, Bourenkov, G. | | Deposit date: | 2017-11-07 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the N-terminal module of the cell wall hydrolase RipA and its role in regulating catalytic activity.
Proteins, 86, 2018
|
|
1YBW
 
 | | Protease domain of HGFA with no inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hepatocyte growth factor activator precursor | | Authors: | Shia, S, Stamos, J, Kirchhofer, D, Fan, B, Wu, J, Corpuz, R.T, Santell, L, Lazarus, R.A, Eigenbrot, C. | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational lability in serine protease active sites: structures of hepatocyte growth factor activator (HGFA) alone and with the inhibitory domain from HGFA inhibitor-1B.
J.Mol.Biol., 346, 2005
|
|
6F0X
 
 | | Cryo-EM structure of TRIP13 in complex with ATP gamma S, p31comet, C-Mad2 and Cdc20 | | Descriptor: | Cell division cycle protein 20 homolog, MAD2L1-binding protein, Mitotic spindle assembly checkpoint protein MAD2A, ... | | Authors: | Alfieri, C, Chang, L, Barford, D. | | Deposit date: | 2017-11-20 | | Release date: | 2018-05-02 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Mechanism for remodelling of the cell cycle checkpoint protein MAD2 by the ATPase TRIP13.
Nature, 559, 2018
|
|
6F0H
 
 | | Crystal structure ASF1-ip4 | | Descriptor: | CITRIC ACID, GLYCEROL, Histone chaperone ASF1A, ... | | Authors: | Bakail, M, Richet, N, Le Du, M.H, Andreani, J, Guerois, R, Ochsenbein, F. | | Deposit date: | 2017-11-20 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Design on a Rational Basis of High-Affinity Peptides Inhibiting the Histone Chaperone ASF1.
Cell Chem Biol, 26, 2019
|
|
6FAP
 
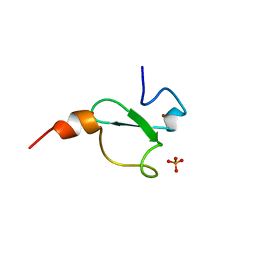 | | Crystal structure of human BAZ2A PHD zinc finger in complex with Fr23 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Amato, A, Lucas, X, Bortoluzzi, A, Wright, D, Ciulli, A. | | Deposit date: | 2017-12-16 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Targeting Ligandable Pockets on Plant Homeodomain (PHD) Zinc Finger Domains by a Fragment-Based Approach.
ACS Chem. Biol., 13, 2018
|
|
1Y98
 
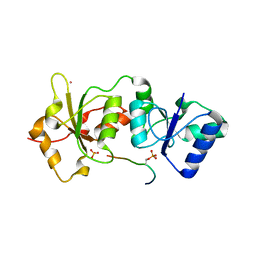 | | Structure of the BRCT repeats of BRCA1 bound to a CtIP phosphopeptide. | | Descriptor: | Breast cancer type 1 susceptibility protein, COBALT (II) ION, CtIP PHOSPHORYLATED PEPTIDE, ... | | Authors: | Varma, A.K, Brown, R.S, Birrane, G, Ladias, J.A.A. | | Deposit date: | 2004-12-14 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Cell Cycle Checkpoint Control by the BRCA1-CtIP Complex.
Biochemistry, 44, 2005
|
|
6FGV
 
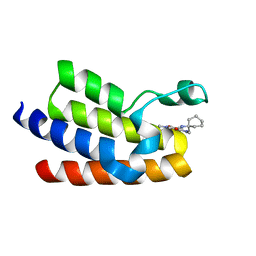 | | Crystal Structure of BAZ2A bromodomain in complex with 1-methylpyridinone compound 3 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, ~{N}-[(1-azanylcyclohexyl)methyl]-1-methyl-6-oxidanylidene-pyridine-3-carboxamide | | Authors: | Dalle Vedove, A, Spiliotopoulos, D, Lolli, G, Caflisch, A. | | Deposit date: | 2018-01-11 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains.
ChemMedChem, 13, 2018
|
|
1UZX
 
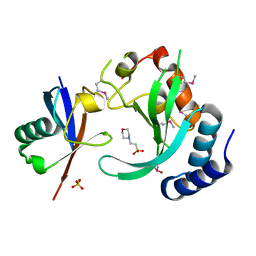 | | A complex of the Vps23 UEV with ubiquitin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, UBIQUITIN, ... | | Authors: | Teo, H, Williams, R.L. | | Deposit date: | 2004-03-18 | | Release date: | 2004-03-30 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights Into Endosomal Sorting Complex Required for Transport (Escrt-I) Recognition of Ubiquitinated Proteins
J.Biol.Chem., 279, 2004
|
|
1XWE
 
 | | NMR Structure of C345C (NTR) domain of C5 of complement | | Descriptor: | Complement C5 | | Authors: | Bramham, J, Thai, C.-T, Soares, D.C, Uhrin, D, Ogata, R.T, Barlow, P.N. | | Deposit date: | 2004-10-30 | | Release date: | 2004-12-21 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Functional Insights from the Structure of the Multifunctional C345C Domain of C5 of Complement
J.Biol.Chem., 280, 2005
|
|
1YX4
 
 | |
1YX6
 
 | |
1ZHB
 
 | | Crystal Structure Of The Murine Class I Major Histocompatibility Complex Of H-2Db, B2-Microglobulin, and a 9-Residue Peptide Derived from rat dopamine beta-monooxigenase | | Descriptor: | 9-mer peptide from Dopamine beta-monooxygenase, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Sandalova, T, Michaelsson, J, Harris, R.A, Odeberg, J, Schneider, G, Karre, K, Achour, A. | | Deposit date: | 2005-04-25 | | Release date: | 2005-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A structural basis for CD8+ T cell-dependent recognition of non-homologous peptide ligands: implications for molecular mimicry in autoreactivity
J.Biol.Chem., 280, 2005
|
|
1YX5
 
 | |
1ZC1
 
 | | Ufd1 exhibits the AAA-ATPase fold with two distinct ubiquitin interaction sites | | Descriptor: | Ubiquitin fusion degradation protein 1 | | Authors: | Park, S, Isaacson, R, Kim, H.T, Silver, P.A, Wagner, G. | | Deposit date: | 2005-04-10 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Ufd1 Exhibits the AAA-ATPase Fold with Two Distinct Ubiquitin Interaction Sites
Structure, 13, 2005
|
|
1ZSG
 
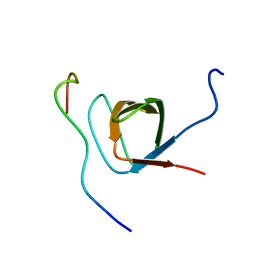 | | beta PIX-SH3 complexed with an atypical peptide from alpha-PAK | | Descriptor: | Rho guanine nucleotide exchange factor 7, Serine/threonine-protein kinase PAK 1 | | Authors: | Mott, H.R, Nietlispach, D, Evetts, K.A, Owen, D. | | Deposit date: | 2005-05-24 | | Release date: | 2005-08-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of the SH3 Domain of beta-PIX and Its Interaction with alpha-p21 Activated Kinase (PAK)
Biochemistry, 44, 2005
|
|
1ZZL
 
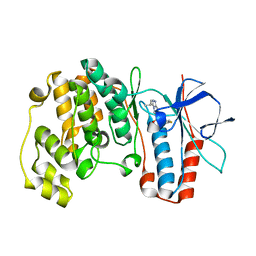 | | Crystal structure of P38 with triazolopyridine | | Descriptor: | 6-[4-(4-FLUOROPHENYL)-1,3-OXAZOL-5-YL]-3-ISOPROPYL[1,2,4]TRIAZOLO[4,3-A]PYRIDINE, Mitogen-activated protein kinase 14 | | Authors: | McClure, K.F, Han, S. | | Deposit date: | 2005-06-14 | | Release date: | 2005-09-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Theoretical and Experimental Design of Atypical Kinase Inhibitors: Application to p38 MAP Kinase.
J.Med.Chem., 48, 2005
|
|
1ZYJ
 
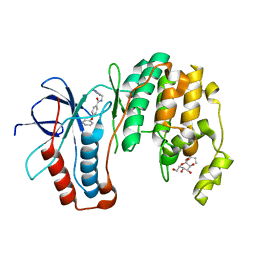 | | Human P38 MAP Kinase in Complex with Inhibitor 1a | | Descriptor: | 4-PHENOXY-N-(PYRIDIN-2-YLMETHYL)BENZAMIDE, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Karpusas, M, Michelotti, E.L, Springman, E.B. | | Deposit date: | 2005-06-10 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two classes of p38alpha MAP kinase inhibitors having a common diphenylether core but exhibiting divergent binding modes.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
3X0W
 
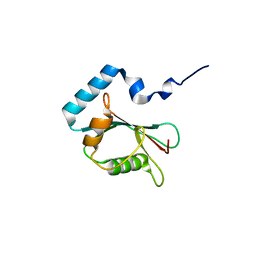 | | Crystal structure of PLEKHM1 LIR-fused human LC3B_2-119 | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Suzuki, H, McEwan, D.G, Popovic, D, Gubas, A, Terawaki, S, Stadel, D, Coxon, F, Stegmann, D.M, Bhogaraju, S, Maddi, K, Kirchhoff, A, Gatti, E, Helfrich, M.H, Behrends, C, Pierre, P, Dikic, I, Wakatsuki, S. | | Deposit date: | 2014-10-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins.
Mol.Cell, 57, 2015
|
|
4UTT
 
 | | Structural characterisation of NanE, ManNac6P C2 epimerase, from Clostridium perfingens | | Descriptor: | ACETATE ION, CHLORIDE ION, PUTATIVE N-ACETYLMANNOSAMINE-6-PHOSPHATE 2-EPIMERASE | | Authors: | Pelissier, M.C, Sebban-Kreuzer, C, Guerlesquin, F, Brannigan, J.A, Davies, G.J, Bourne, Y, Vincent, F. | | Deposit date: | 2014-07-23 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural and Functional Characterization of the Clostridium Perfringens N-Acetylmannosamine-6-Phosphate 2-Epimerase Essential for the Sialic Acid Salvage Pathway.
J.Biol.Chem., 289, 2014
|
|
5MDU
 
 | | Structure of the RNA recognition motif (RRM) of Seb1 from S. pombe. | | Descriptor: | CHLORIDE ION, GLYCEROL, Rpb7-binding protein seb1, ... | | Authors: | Wittmann, S, Renner, M, El Omari, K, Adams, O, Vasiljeva, L, Grimes, J. | | Deposit date: | 2016-11-13 | | Release date: | 2017-04-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | The conserved protein Seb1 drives transcription termination by binding RNA polymerase II and nascent RNA.
Nat Commun, 8, 2017
|
|
4V3R
 
 | | Designed armadillo repeat protein with 5 internal repeats, 2nd generation C-cap and 3rd generation N-cap. | | Descriptor: | MAGNESIUM ION, YIII_M5_AII | | Authors: | Reichen, C, Madhurantakam, C, Pluckthun, A, Mittl, P. | | Deposit date: | 2014-10-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of Designed Armadillo-Repeat Proteins Show Propagation of Inter-Repeat Interface Effects
Acta Crystallogr.,Sect.D, 72, 2016
|
|
5MPY
 
 | | Crystal structure of Arabidopsis thaliana RNA editing factor MORF9 | | Descriptor: | CALCIUM ION, Multiple organellar RNA editing factor 9, chloroplastic | | Authors: | Haag, S, Schindler, M, Berndt, L, Brennicke, A, Takenaka, M, Weber, G. | | Deposit date: | 2016-12-19 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | Crystal structures of the Arabidopsis thaliana organellar RNA editing factors MORF1 and MORF9.
Nucleic Acids Res., 45, 2017
|
|
4V28
 
 | | Structure of an E333Q variant of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with Man-Man-Methylumbelliferone | | Descriptor: | 1,2-ETHANEDIOL, 7-hydroxy-4-methyl-2H-chromen-2-one, ACETATE ION, ... | | Authors: | Hakki, Z, Bellmaine, S, Thompson, A.J, Speciale, G, Davies, G.J, Williams, S.J. | | Deposit date: | 2014-10-07 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and Kinetic Dissection of the Endo-Alpha-1,2-Mannanase Activity of Bacterial Gh99 Glycoside Hydrolases from Bacteroides Spp.
Chemistry, 21, 2015
|
|
5MGK
 
 | |
