3K1T
 
 | |
1V8B
 
 | | Crystal structure of a hydrolase | | Descriptor: | ADENOSINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, adenosylhomocysteinase | | Authors: | Tanaka, N, Nakanishi, M, Kusakabe, Y, Shiraiwa, K, Kitade, Y, Nakamura, K.T. | | Deposit date: | 2004-01-03 | | Release date: | 2004-10-26 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of S-Adenosyl-l-Homocysteine Hydrolase from the Human Malaria Parasite Plasmodium falciparum
J.Mol.Biol., 343, 2004
|
|
1UMY
 
 | | BHMT from rat liver | | Descriptor: | BETA-MERCAPTOETHANOL, Betaine--homocysteine S-methyltransferase 1, ZINC ION | | Authors: | Gonzalez, B, Pajares, M.A, Sanz-Aparicio, J. | | Deposit date: | 2003-09-02 | | Release date: | 2004-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of rat liver betaine homocysteine s-methyltransferase reveals new oligomerization features and conformational changes upon substrate binding.
J. Mol. Biol., 338, 2004
|
|
1GR7
 
 | | Crystal structure of the double mutant Cys3Ser/Ser100Pro from Pseudomonas Aeruginosa at 1.8 A resolution | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Okvist, M, Bonander, N, Sandberg, A, Karlsson, B.G, Krengel, U, Xue, Y, Sjolin, L. | | Deposit date: | 2001-12-14 | | Release date: | 2002-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the double azurin mutant Cys3Ser/Ser100Pro from Pseudomonas aeruginosa at 1.8 A resolution: its folding-unfolding energy and unfolding kinetics.
Biochim.Biophys.Acta, 1596, 2002
|
|
7BZL
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with beta-L-arabinofuranose-configured cyclophellitol | | Descriptor: | (1S,2S,3R,4R)-3-(hydroxymethyl)cyclopentane-1,2,4-triol, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Amaki, S, McGregor, N.G.S, Arakawa, T, Yamada, C, Borlandelli, V, Overkleeft, H.S, Davies, G.J, Fushinobu, S. | | Deposit date: | 2020-04-28 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cysteine Nucleophiles in Glycosidase Catalysis: Application of a Covalent beta-l-Arabinofuranosidase Inhibitor.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DIF
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with beta-L-arabinofuranose-configured cyclophellitol at 1.75-angstrom resolution | | Descriptor: | (1S,2S,3R,4R)-3-(hydroxymethyl)cyclopentane-1,2,4-triol, Non-reducing end beta-L-arabinofuranosidase, POTASSIUM ION, ... | | Authors: | Amaki, S, McGregor, N.G.S, Arakawa, T, Yamada, C, Borlandelli, V, Overkleeft, H.S, Davies, G.J, Fushinobu, S. | | Deposit date: | 2020-11-19 | | Release date: | 2021-01-27 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cysteine Nucleophiles in Glycosidase Catalysis: Application of a Covalent beta-l-Arabinofuranosidase Inhibitor.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
1BBO
 
 | |
1PYI
 
 | |
2DCW
 
 | |
2DCV
 
 | |
9M0Q
 
 | |
2I4N
 
 | | Rhodopseudomonas palustris prolyl-tRNA synthetase in complex with CysAMS | | Descriptor: | 5'-O-(N-(L-CYSTEINYL)-SULFAMOYL)ADENOSINE, Proline-tRNA ligase | | Authors: | Crepin, T, Yaremchuk, A, Tukalo, M, Cusack, S. | | Deposit date: | 2006-08-22 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of Two Bacterial Prolyl-tRNA Synthetases with and without a cis-Editing Domain.
Structure, 14, 2006
|
|
4U84
 
 | | Human Pin1 with S-hydroxyl-cysteine 113 | | Descriptor: | POLYETHYLENE GLYCOL (N=34), Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Li, W, Zhang, Y. | | Deposit date: | 2014-08-01 | | Release date: | 2015-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Pin1 cysteine-113 oxidation inhibits its catalytic activity and cellular function in Alzheimer's disease.
Neurobiol.Dis., 76, 2015
|
|
1G2F
 
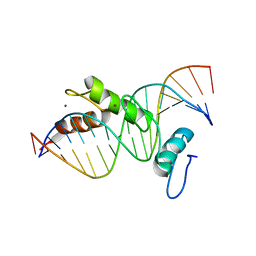 | | STRUCTURE OF A CYS2HIS2 ZINC FINGER/TATA BOX COMPLEX (TATAZF;CLONE #6) | | Descriptor: | 5'-D(*GP*AP*CP*GP*CP*TP*AP*TP*AP*AP*AP*AP*GP*GP*AP*G)-3', 5'-D(*TP*CP*CP*TP*TP*TP*TP*AP*TP*AP*GP*CP*GP*TP*CP*C)-3', TATA BOX ZINC FINGER PROTEIN, ... | | Authors: | Wolfe, S.A, Grant, R.A, Elrod-Erickson, M, Pabo, C.O. | | Deposit date: | 2000-10-18 | | Release date: | 2001-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Beyond the "recognition code": structures of two Cys2His2 zinc finger/TATA box complexes.
Structure, 9, 2001
|
|
1G2D
 
 | | STRUCTURE OF A CYS2HIS2 ZINC FINGER/TATA BOX COMPLEX (CLONE #2) | | Descriptor: | 5'-D(*GP*AP*CP*GP*CP*TP*AP*TP*AP*AP*AP*AP*GP*GP*AP*G)-3', 5'-D(*TP*CP*CP*TP*TP*TP*TP*AP*TP*AP*GP*CP*GP*TP*CP*C)-3', TATA BOX ZINC FINGER PROTEIN, ... | | Authors: | Wolfe, S.A, Grant, R.A, Elrod-Erickson, M, Pabo, C.O. | | Deposit date: | 2000-10-18 | | Release date: | 2001-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Beyond the "recognition code": structures of two Cys2His2 zinc finger/TATA box complexes.
Structure, 9, 2001
|
|
8S03
 
 | | NMR solution structure of the CysD2 domain of MUC2 | | Descriptor: | CALCIUM ION, Mucin-2 | | Authors: | Recktenwald, C, Karlsson, B.G, Garcia-Bonnete, M.-J, Katona, G, Jensen, M, Lymer, R, Baeckstroem, M, Johansson, M.E.V, Hansson, G.C, Trillo-Muyo, S. | | Deposit date: | 2024-02-13 | | Release date: | 2024-04-17 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | The structure of the second CysD domain of MUC2 and role in mucin organization by transglutaminase-based cross-linking.
Cell Rep, 43, 2024
|
|
2B1M
 
 | | Crystal structure of a papain-fold protein without the catalytic cysteine from seeds of Pachyrhizus erosus | | Descriptor: | DI(HYDROXYETHYL)ETHER, SPE31, TETRAETHYLENE GLYCOL, ... | | Authors: | Zhang, M, Wei, Z, Chang, S. | | Deposit date: | 2005-09-16 | | Release date: | 2006-10-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a papain-fold protein without the catalytic residue: a novel member in the cysteine proteinase family
J.Mol.Biol., 358, 2006
|
|
2B1N
 
 | |
6ELW
 
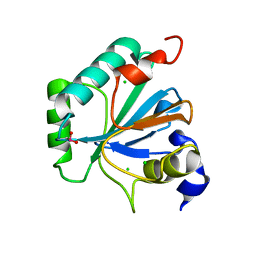 | | High resolution structure of selenocysteine containing human GPX4 | | Descriptor: | CHLORIDE ION, Phospholipid hydroperoxide glutathione peroxidase, mitochondrial | | Authors: | Kalms, J, Borchert, A, Kuhn, H, Scheerer, P. | | Deposit date: | 2017-09-29 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure and functional characterization of selenocysteine-containing glutathione peroxidase 4 suggests an alternative mechanism of peroxide reduction.
Biochim. Biophys. Acta, 1863, 2018
|
|
8ZKS
 
 | |
8ZKH
 
 | |
8ZKT
 
 | |
8ZKR
 
 | |
8ZKU
 
 | |
6YQH
 
 | | GH146 beta-L-arabinofuranosidase bound to covalent inhibitor | | Descriptor: | (1~{S},2~{S},3~{S},4~{S})-4-(hydroxymethyl)cyclopentane-1,2,3-triol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acetyl-CoA carboxylase, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-04-17 | | Release date: | 2021-01-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Cysteine Nucleophiles in Glycosidase Catalysis: Application of a Covalent beta-l-Arabinofuranosidase Inhibitor.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
