2ZNT
 
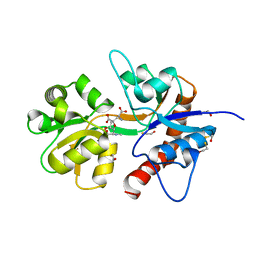 | | Crystal structure of the ligand-binding core of the human ionotropic glutamate receptor, GluR5, in complex with a novel selective agonist, dysiherbaine | | Descriptor: | (2R,3aR,6S,7R,7aR)-2-[(2S)-2-amino-2-carboxyethyl]-6-hydroxy-7-(methylamino)hexahydro-2H-furo[3,2-b]pyran-2-carboxylic acid, BETA-MERCAPTOETHANOL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2008-05-01 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
2YV9
 
 | |
2YKR
 
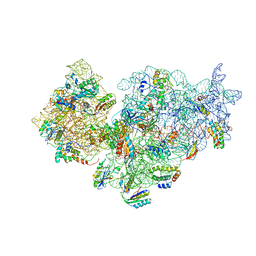 | | 30S ribosomal subunit with RsgA bound in the presence of GMPPNP | | Descriptor: | 16S RRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Guo, Q, Yuan, Y, Xu, Y, Feng, B, Liu, L, Chen, K, Lei, J, Gao, N. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Structural Basis for the Function of a Small Gtpase Rsga on the 30S Ribosomal Subunit Maturation Revealed by Cryoelectron Microscopy.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1MAA
 
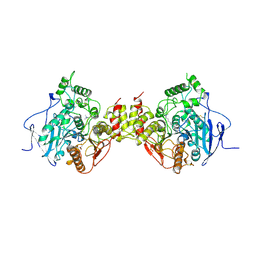 | | MOUSE ACETYLCHOLINESTERASE CATALYTIC DOMAIN, GLYCOSYLATED PROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Bourne, Y, Taylor, P, Bougis, P.E, Marchot, P. | | Deposit date: | 1998-11-04 | | Release date: | 1999-04-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of mouse acetylcholinesterase. A peripheral site-occluding loop in a tetrameric assembly.
J.Biol.Chem., 274, 1999
|
|
1K40
 
 | |
1KC4
 
 | | NMR Structural Analysis of the Complex Formed Between alpha-Bungarotoxin and the Principal alpha-Neurotoxin Binding Sequence on the alpha7 Subunit of a Neuronal Nicotinic Acetylcholine Receptor | | Descriptor: | alpha-bungarotoxin, neuronal acetylcholine receptor protein, alpha-7 chain | | Authors: | Moise, L, Piserchio, A, Basus, V.J, Hawrot, E. | | Deposit date: | 2001-11-07 | | Release date: | 2002-03-13 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | NMR structural analysis of alpha-bungarotoxin and its complex with the principal alpha-neurotoxin-binding sequence on the alpha 7 subunit of a neuronal nicotinic acetylcholine receptor.
J.Biol.Chem., 277, 2002
|
|
2YFH
 
 | | Structure of a Chimeric Glutamate Dehydrogenase | | Descriptor: | GLUTAMATE DEHYDROGENASE, NAD-SPECIFIC GLUTAMATE DEHYDROGENASE | | Authors: | Oliveira, T, Panjikar, S, Sharkey, M.A, Carrigan, J.B, Hamza, M, Engel, P.C, Khan, A.R. | | Deposit date: | 2011-04-05 | | Release date: | 2012-04-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Crystal Structure of a Chimaeric Bacterial Glutamate Dehydrogenase.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
1KL8
 
 | | NMR STRUCTURAL ANALYSIS OF THE COMPLEX FORMED BETWEEN ALPHA-BUNGAROTOXIN AND THE PRINCIPAL ALPHA-NEUROTOXIN BINDING SEQUENCE ON THE ALPHA7 SUBUNIT OF A NEURONAL NICOTINIC ACETYLCHOLINE RECEPTOR | | Descriptor: | ALPHA-BUNGAROTOXIN, NEURONAL ACETYLCHOLINE RECEPTOR PROTEIN, ALPHA-7 CHAIN | | Authors: | Moise, L, Piserchio, A, Basus, V.J, Hawrot, E. | | Deposit date: | 2001-12-11 | | Release date: | 2002-03-13 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | NMR structural analysis of alpha-bungarotoxin and its complex with the principal alpha-neurotoxin-binding sequence on the alpha 7 subunit of a neuronal nicotinic acetylcholine receptor.
J.Biol.Chem., 277, 2002
|
|
1IHC
 
 | | X-ray Structure of Gephyrin N-terminal Domain | | Descriptor: | Gephyrin | | Authors: | Sola, M, Kneussel, M, Heck, I.S, Betz, H, Weissenhorn, W. | | Deposit date: | 2001-04-21 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystal structure of the trimeric N-terminal domain of gephyrin.
J.Biol.Chem., 276, 2001
|
|
1ITU
 
 | | HUMAN RENAL DIPEPTIDASE COMPLEXED WITH CILASTATIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CILASTATIN, RENAL DIPEPTIDASE, ... | | Authors: | Nitanai, Y, Satow, Y, Adachi, H, Tsujimoto, M. | | Deposit date: | 2002-02-03 | | Release date: | 2002-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Renal Dipeptidase Involved in beta-Lactam Hydrolysis
J.Mol.Biol., 321, 2002
|
|
1IDH
 
 | | THE NMR SOLUTION STRUCTURE OF THE COMPLEX FORMED BETWEEN ALPHA-BUNGAROTOXIN AND AN 18MER COGNATE PEPTIDE | | Descriptor: | ACETYLCHOLINE RECEPTOR PROTEIN, ALPHA CHAIN, ALPHA-BUNGAROTOXIN | | Authors: | Zeng, H, Moise, L, Grant, M.A, Hawrot, E. | | Deposit date: | 2001-04-04 | | Release date: | 2001-04-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the complex formed between alpha-bungarotoxin and an 18-mer cognate peptide derived from the alpha 1 subunit of the nicotinic acetylcholine receptor from Torpedo californica.
J.Biol.Chem., 276, 2001
|
|
3AG3
 
 | | Bovine Heart Cytochrome c Oxidase in the Nitric Oxide-bound Fully Reduced State at 100 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Muramoto, K, Ohta, K, Shinzawa-Itoh, K, Kanda, K, Taniguchi, M, Nabekura, H, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2010-03-19 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1IM3
 
 | | Crystal Structure of the human cytomegalovirus protein US2 bound to the MHC class I molecule HLA-A2/tax | | Descriptor: | HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, A-2 ALPHA CHAIN, Human T-cell lymphotropic virus type 1 Tax peptide, ... | | Authors: | Gewurz, B.E, Gaudet, R, Tortorella, D, Wang, E.W, Ploegh, H.L, Wiley, D.C. | | Deposit date: | 2001-05-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antigen presentation subverted: Structure of the human cytomegalovirus protein US2 bound to the class I molecule HLA-A2.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1ORV
 
 | | Crystal Structure of Porcine Dipeptidyl Peptidase IV (CD26) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | Authors: | Engel, M, Hoffmann, T, Wagner, L, Wermann, M, Heiser, U, Kiefersauer, R, Huber, R, Bode, W, Demuth, H.U, Brandstetter, H. | | Deposit date: | 2003-03-16 | | Release date: | 2003-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of Dipeptidyl Peptidase IV (CD26) Reveals its Functional Regulation and Enzymatic Mechanism
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1OW8
 
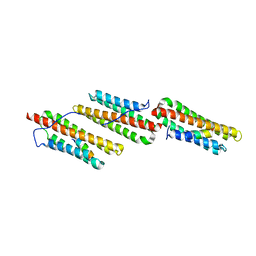 | | Paxillin LD2 motif bound to the Focal Adhesion Targeting (FAT) domain of the Focal Adhesion Kinase | | Descriptor: | Focal adhesion kinase 1, Paxillin | | Authors: | Hoellerer, M.K, Noble, M.E.M, Labesse, G, Werner, J.M, Arold, S.T. | | Deposit date: | 2003-03-28 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular Recognition of Paxillin LD Motifs
by the Focal Adhesion Targeting Domain
Structure, 11, 2003
|
|
1MP8
 
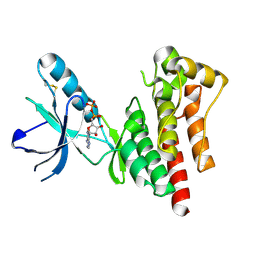 | | Crystal structure of Focal Adhesion Kinase (FAK) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, focal adhesion kinase 1 | | Authors: | Nowakowski, J, Cronin, C.N, McRee, D.E, Knuth, M.W, Nelson, C.G, Pavletich, N.P, Rodgers, J, Sang, B.-C, Scheibe, D.N, Swanson, R.V, Thompson, D.A. | | Deposit date: | 2002-09-11 | | Release date: | 2003-09-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of the cancer-related Aurora-A, FAK, and EphA2 protein kinases from nanovolume crystallography
Structure, 10, 2002
|
|
1G0Y
 
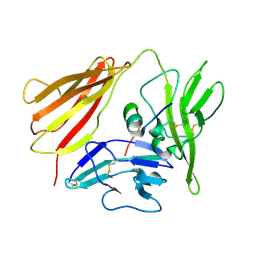 | | IL-1 RECEPTOR TYPE 1 COMPLEXED WITH ANTAGONIST PEPTIDE AF10847 | | Descriptor: | ANTAGONIST PEPTIDE AF10847, INTERLEUKIN-1 RECEPTOR, TYPE I | | Authors: | Vigers, G.P.A, Dripps, D.J, Edwards, C.K, Brandhuber, B.J. | | Deposit date: | 2000-10-09 | | Release date: | 2000-10-25 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray crystal structure of a small antagonist peptide bound to interleukin-1 receptor type 1.
J.Biol.Chem., 275, 2000
|
|
1KFT
 
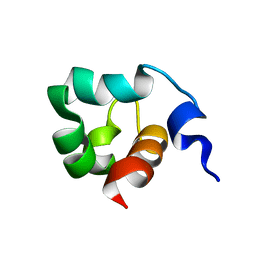 | | Solution Structure of the C-Terminal domain of UvrC from E-coli | | Descriptor: | Excinuclease ABC subunit C | | Authors: | Singh, S, Folkers, G.E, Bonvin, A.M.J.J, Boelens, R, Wechselberger, R, Niztayev, A, Kaptein, R. | | Deposit date: | 2001-11-23 | | Release date: | 2002-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and DNA-binding properties of the C-terminal domain of UvrC from E.coli
EMBO J., 21, 2002
|
|
1K04
 
 | |
1LXH
 
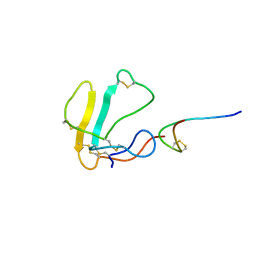 | |
3A5L
 
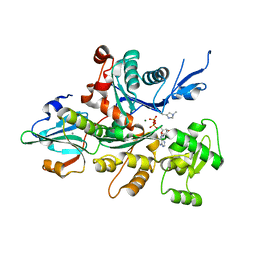 | | Crystal Structure of a Dictyostelium P109A Mg2+-Actin in Complex with Human Gelsolin Segment 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Gelsolin, ... | | Authors: | Murakami, K, Yasunaga, T, Noguchi, T.Q, Uyeda, T.Q, Wakabayashi, T. | | Deposit date: | 2009-08-09 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release
Cell(Cambridge,Mass.), 143, 2010
|
|
1MZC
 
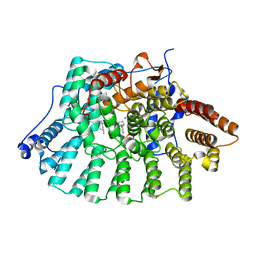 | | Co-Crystal Structure Of Human Farnesyltransferase With Farnesyldiphosphate and Inhibitor Compound 33a | | Descriptor: | 2-[3-(3-ETHYL-1-METHYL-2-OXO-AZEPAN-3-YL)-PHENOXY]-4-[1-AMINO-1-(1-METHYL-1H-IMIDIZOL-5-YL)-ETHYL]-BENZONITRILE, FARNESYL DIPHOSPHATE, Protein Farnesyltransferase alpha Subunit, ... | | Authors: | deSolms, S.J, Ciccarone, T.M, MacTough, S.C, Shaw, A.W, Buser, C.A, Ellis-Hutchings, M, Fernandes, C, Hamilton, K.A, Huber, H.E, Kohl, N.E, Lobell, R.B, Robinson, R.G, Tsou, N.N, Walsh, E.S, Graham, S.L, Beese, L.S, Taylor, J.S. | | Deposit date: | 2002-10-07 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dual Protein Farnesyltransferase-Geranylgeranyltransferase-I Inhibitors as Potential Cancer Chemotherapeutic Agents.
J.Med.Chem., 46, 2003
|
|
1MQZ
 
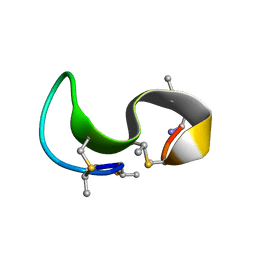 | | NMR solution structure of type-B lantibiotics mersacidin bound to lipid II in DPC micelles | | Descriptor: | LANTIBIOTIC MERSACIDIN | | Authors: | Hsu, S.-T, Breukink, E, Bierbaum, G, Sahl, H.-G, de Kruijff, B, Kaptein, R, van Nuland, N.A, Bonvin, A.M. | | Deposit date: | 2002-09-17 | | Release date: | 2003-03-11 | | Last modified: | 2024-07-10 | | Method: | SOLUTION NMR | | Cite: | NMR Study of Mersacidin and Lipid II Interaction in Dodecylphosphocholine Micelles. Conformational Changes are a Key to Antimicrobial Activity
J.Biol.Chem., 278, 2003
|
|
3AG4
 
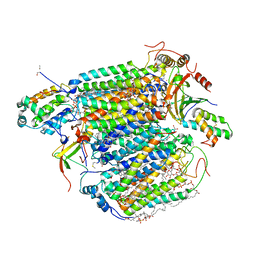 | | Bovine Heart Cytochrome c Oxidase in the Cyanide Ion-bound Fully Reduced State at 100 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Muramoto, K, Ohta, K, Shinzawa-Itoh, K, Kanda, K, Taniguchi, M, Nabekura, H, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2010-03-19 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3A5O
 
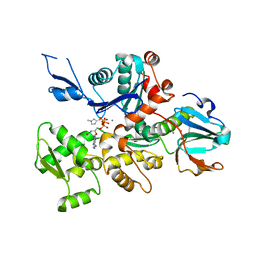 | | Crystal Structure of a Dictyostelium P109I Ca2+-Actin in Complex with Human Gelsolin Segment 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Gelsolin, ... | | Authors: | Murakami, K, Yasunaga, T, Noguchi, T.Q, Uyeda, T.Q, Wakabayashi, T. | | Deposit date: | 2009-08-09 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release
Cell(Cambridge,Mass.), 143, 2010
|
|
