8OU1
 
 | |
2RSJ
 
 | |
2RSH
 
 | |
1KQJ
 
 | | Crystal Structure of a Mutant of MutY Catalytic Domain | | Descriptor: | A/G-SPECIFIC ADENINE GLYCOSYLASE, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Messick, T.E, Chmiel, N.H, Golinelli, M.P, David, S.S, Joshua-Tor, L. | | Deposit date: | 2002-01-06 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Noncysteinyl coordination to the [4Fe-4S]2+ cluster of the DNA repair adenine glycosylase MutY introduced via site-directed mutagenesis. Structural characterization of an unusual histidinyl-coordinated cluster.
Biochemistry, 41, 2002
|
|
4X18
 
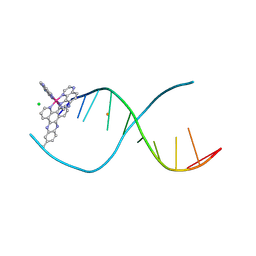 | | [Ru(TAP)2(dppz-11-Me)]2+ bound to d(TCGGCGCCGA) | | Descriptor: | (11-methyldipyrido[3,2-a:2',3'-c]phenazine-kappa~2~N~4~,N~5~)[bis(pyrazino[2,3-f]quinoxaline-kappa~2~N~1~,N~10~)]ruthenium, BARIUM ION, CHLORIDE ION, ... | | Authors: | Hall, J.P, Cardin, C.J. | | Deposit date: | 2014-11-24 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The Structural Effect of Methyl Substitution on the Binding of Polypyridyl Ru-dppz Complexes to DNA
Organometallics, 2015
|
|
4X1A
 
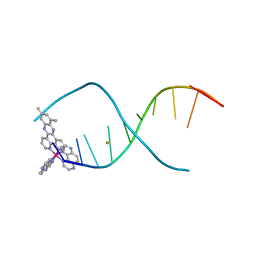 | | Lambda-[Ru(TAP)2(dppz-10,12-Me)]2+ bound to d(TCGGCGCCGA) | | Descriptor: | (10,12-dimethyldipyrido[3,2-a:2',3'-c]phenazine-kappa~2~N~4~,N~5~)[bis(pyrazino[2,3-f]quinoxaline-kappa~2~N~1~,N~10~)]ruthenium, BARIUM ION, DNA (5'-D(*TP*CP*GP*GP*CP*GP*CP*CP*GP*A)-3') | | Authors: | Hall, J.P, Cardin, C.J. | | Deposit date: | 2014-11-24 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | The Structural Effect of Methyl Substitution on the Binding of Polypyridyl Ru-dppz Complexes to DNA
Organometallics, 2015
|
|
8DFU
 
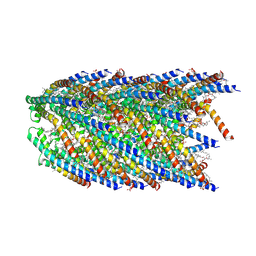 | | Cryo-EM structure of conjugation pili from Aeropyrum pernix | | Descriptor: | (2S)-3-{[(3R,7S,11S,15S)-3,7,11,15,19-pentamethylicosyl]oxy}-2-{[(2R,6S,10S,14R)-2,6,10,14,18-pentamethylnonadecyl]oxy}propyl dihydrogen phosphate, Pilin protein | | Authors: | Beltran, L.C, Egelman, E.H. | | Deposit date: | 2022-06-22 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Archaeal DNA-import apparatus is homologous to bacterial conjugation machinery.
Nat Commun, 14, 2023
|
|
8DFT
 
 | | Cryo-EM structure of conjugative pili from Pyrobaculum calidifontis | | Descriptor: | Pilin protein, [(2~{S},7~{S},11~{S},15~{S},19~{R},22~{R},26~{S},30~{R},34~{R},38~{S},43~{S},47~{S},51~{S},55~{R},58~{R},62~{S},66~{R},70~{R})-38-(hydroxymethyl)-7,11,15,19,22,26,30,34,43,47,51,55,58,62,66,70-hexadecamethyl-1,4,37,40-tetraoxacyclodoheptacont-2-yl]methanol | | Authors: | Beltran, L.C, Egelman, E.H. | | Deposit date: | 2022-06-22 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Archaeal DNA-import apparatus is homologous to bacterial conjugation machinery.
Nat Commun, 14, 2023
|
|
1G80
 
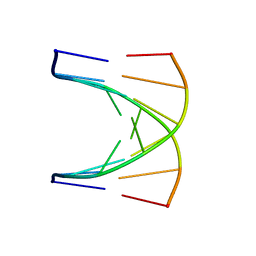 | | NMR SOLUTION STRUCTURE OF D(GCGTACGC)2 | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*CP*GP*C)-3' | | Authors: | Isaacs, R.J, Spielmann, H.P. | | Deposit date: | 2000-11-15 | | Release date: | 2001-03-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Relationship of DNA structure to internal dynamics: correlation of helical parameters from NOE-based NMR solution structures of d(GCGTACGC)(2) and d(CGCTAGCG)(2) with (13)C order parameters implies conformational coupling in dinucleotide units.
J.Mol.Biol., 307, 2001
|
|
7VIM
 
 | |
6QXM
 
 | | Cryo-EM structure of T7 bacteriophage portal protein, 12mer, open valve | | Descriptor: | Portal protein | | Authors: | Fabrega-Ferrer, M, Cuervo, A, Machon, C, Fernandez, F.J, Perez-Luque, R, Pous, J, Vega, M.C, Carrascosa, J.L, Coll, M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of T7 bacteriophage portal and tail suggest a viral DNA retention and ejection mechanism.
Nat Commun, 10, 2019
|
|
6QWP
 
 | | Crystal structure of T7 bacteriophage portal protein, 13mer, closed valve | | Descriptor: | Portal protein | | Authors: | Fabrega-Ferrer, M, Cuervo, A, Machon, C, Fernandez, F.J, Perez-Luque, R, Pous, J, Vega, M.C, Carrascosa, J.L, Coll, M. | | Deposit date: | 2019-03-06 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of T7 bacteriophage portal and tail suggest a viral DNA retention and ejection mechanism.
Nat Commun, 10, 2019
|
|
7VYX
 
 | | Crystal structure of the selenomethionine(SeMet)-derived Cas12c1 (D969A) ternary complex | | Descriptor: | Non-target DNA strand, Selenomethionine (SeMet)-labeled Cas12c1 D969A mutant, Target DNA strand, ... | | Authors: | Zhang, B, Lin, J.Y, Perculija, V, OuYang, S.Y. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-02 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into target DNA recognition and cleavage by the CRISPR-Cas12c1 system
Nucleic Acids Res., 50, 2022
|
|
8EB5
 
 | |
6TX1
 
 | | HPF1 from Nematostella vectensis | | Descriptor: | Predicted protein | | Authors: | Suskiewicz, M.J, Ahel, I. | | Deposit date: | 2020-01-13 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | HPF1 completes the PARP active site for DNA damage-induced ADP-ribosylation.
Nature, 579, 2020
|
|
6TVH
 
 | |
1G7Z
 
 | | NMR SOLUTION STRUCTURE OF D(CGCTAGCG)2 | | Descriptor: | 5'-D(*CP*GP*CP*TP*AP*GP*CP*G)-3' | | Authors: | Isaacs, R.J, Spielmann, H.P. | | Deposit date: | 2000-11-15 | | Release date: | 2001-03-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Relationship of DNA structure to internal dynamics: correlation of helical parameters from NOE-based NMR solution structures of d(GCGTACGC)(2) and d(CGCTAGCG)(2) with (13)C order parameters implies conformational coupling in dinucleotide units.
J.Mol.Biol., 307, 2001
|
|
1ZEZ
 
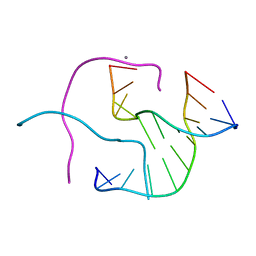 | | ACC Holliday Junction | | Descriptor: | 5'-D(*CP*CP*GP*GP*TP*AP*CP*CP*GP*G)-3', CALCIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-19 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
7FB7
 
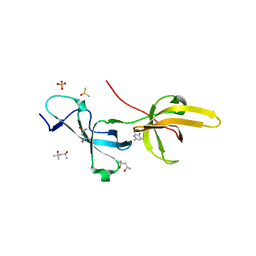 | | Crystal structure of human UHRF1 TTD in complex with 5-amino-2,4-dimethylpyridine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-amino-2,4-dimethylpyridine, DIMETHYL SULFOXIDE, ... | | Authors: | Kori, S, Arita, K, Yoshimi, S. | | Deposit date: | 2021-07-08 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-based screening combined with computational and biochemical analyses identified the inhibitor targeting the binding of DNA Ligase 1 to UHRF1.
Bioorg.Med.Chem., 52, 2021
|
|
5J7O
 
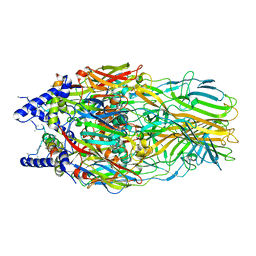 | | Faustovirus major capsid protein | | Descriptor: | major capsid protein, unknown | | Authors: | Klose, T, Rossmann, M.G. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure of faustovirus, a large dsDNA virus.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5J7U
 
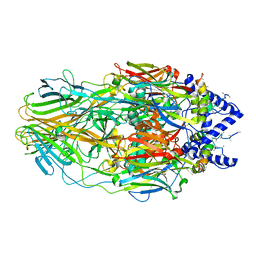 | | Faustovirus major capsid protein | | Descriptor: | major capsid protein, unknown | | Authors: | Klose, T, Rossmann, M.G. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structure of faustovirus, a large dsDNA virus.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1NTQ
 
 | | 5'(dCCUCCUU)3':3'(rAGGAGGAAA)5' | | Descriptor: | 5'-D(*CP*CP*UP*CP*CP*UP*U)-3', 5'-R(*AP*AP*AP*GP*GP*AP*GP*GP*A)-3' | | Authors: | Znosko, B.M, Barnes III, T.W, Krugh, T.R, Turner, D.H. | | Deposit date: | 2003-01-30 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Studies of DNA Single Strands and DNA:RNA Hybrids With and Without 1-Propynylation at C5 of Oligopyrimidines
J.Am.Chem.Soc., 125, 2003
|
|
5J7V
 
 | | Faustovirus major capsid protein | | Descriptor: | major capsid protein | | Authors: | Klose, T, Rossmann, M.G. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (15.5 Å) | | Cite: | Structure of faustovirus, a large dsDNA virus.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7D8K
 
 | | Solution structure of the methyl-CpG binding domain of MBD6 from Arabidopsis thaliana | | Descriptor: | Methyl-CpG-binding domain-containing protein 6 | | Authors: | Mahana, Y, Ohki, I, Walinda, E, Morimoto, D, Sugase, K, Shirakawa, M. | | Deposit date: | 2020-10-08 | | Release date: | 2021-10-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Insights into Methylated DNA Recognition by the Methyl-CpG Binding Domain of MBD6 from Arabidopsis thaliana .
Acs Omega, 7, 2022
|
|
2WQ6
 
 | | Structure of the 6-4 photolyase of D. melanogaster in complex with the non-natural N4-methyl T(Dewar)C lesion | | Descriptor: | 5'-D(*AP*CP*AP*GP*CP*GP*GP*TDYP*CDWP*GP* CP*AP*AP*GP*T)-3', 5'-D(*TP*AP*CP*CP*TP*GP*CP*GP*AP*CP* CP*GP*CP*TP*G)-3', FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Glas, A.F, Kaya, E, Schneider, S, Maul, M.J, Carell, T. | | Deposit date: | 2009-08-14 | | Release date: | 2010-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA (6-4) Photolyases Reduce Dewar Isomers for Isomerization Into (6-4) Lesions
J.Am.Chem.Soc., 132, 2010
|
|
