6WAJ
 
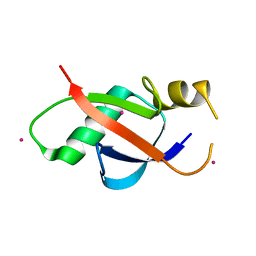 | | Crystal structure of the UBL domain of human NLE1 | | Descriptor: | NLE1, UNKNOWN ATOM OR ION | | Authors: | Halabelian, L, Zeng, H, Li, Y, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-25 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the UBL domain of human NLE1
To be Published
|
|
5L6W
 
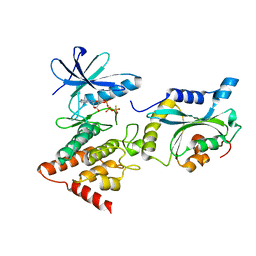 | | Structure Of the LIMK1-ATPgammaS-CFL1 Complex | | Descriptor: | Cofilin-1, LIM domain kinase 1, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Salah, E, Mathea, S, Oerum, S, Newman, J.A, Tallant, C, Adamson, R, Canning, P, Beltrami, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Bullock, A.N. | | Deposit date: | 2016-05-31 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure Of the LIMK1-ATPgammaS-CFL1 Complex
To Be Published
|
|
6WAN
 
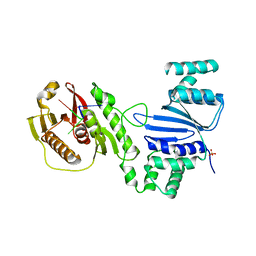 | | Structure of Acinetobacter baumannii Cap4 SAVED/CARF-domain containing receptor with the cyclic trinucleotide 3'3'3'-cAAA | | Descriptor: | Cyclic RNA (R(P*AP*AP*A), SAVED domain-containing protein, SULFATE ION | | Authors: | Lowey, B, Whiteley, A.T, Keszei, A.F.A, Morehouse, B.R, Antine, S.P, Cabrera, V, Schwede, F, Mekalanos, J.J, Shao, S, Lee, A.S.Y, Kranzusch, P.J. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CBASS Immunity Uses CARF-Related Effectors to Sense 3'-5'- and 2'-5'-Linked Cyclic Oligonucleotide Signals and Protect Bacteria from Phage Infection.
Cell, 182, 2020
|
|
4YLZ
 
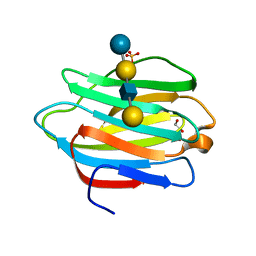 | |
8FV0
 
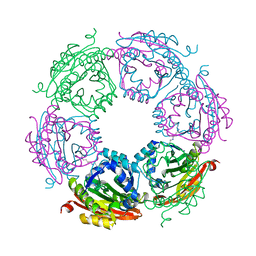 | |
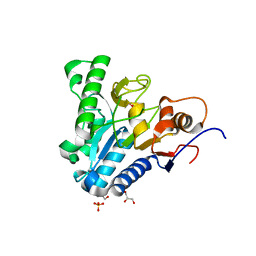
6VYE
 
 | |
5L7I
 
 | | Structure of human Smoothened in complex with Vismodegib | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-chloranyl-~{N}-(4-chloranyl-3-pyridin-2-yl-phenyl)-4-methylsulfonyl-benzamide, SODIUM ION, ... | | Authors: | Byrne, E.X.B, Sircar, R, Miller, P.S, Hedger, G, Luchetti, G, Nachtergaele, S, Tully, M.D, Mydock-McGrane, L, Covey, D.F, Rambo, R.F, Sansom, M.S.P, Newstead, S, Rohatgi, R, Siebold, C. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of Smoothened regulation by its extracellular domains.
Nature, 535, 2016
|
|
8FHJ
 
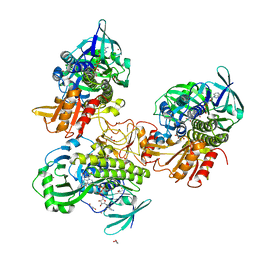 | | Crystal structure of a FAD monooxygenease from Methylocystis sp. Strain SB2 | | Descriptor: | BROMIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Stewart, A.M, Sawaya, M.R, Stewart, C.E. | | Deposit date: | 2022-12-14 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of MbnF: an NADPH-dependent flavin monooxygenase from Methylocystis strain SB2.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
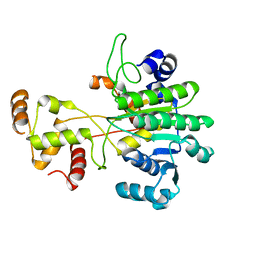
6VTZ
 
 | |
5L8C
 
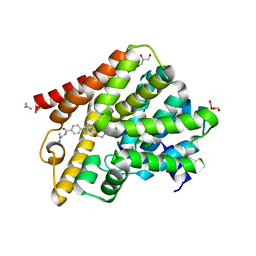 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-039 | | Descriptor: | 4-[5-[(4~{a}~{R},8~{a}~{S})-4-oxidanylidene-3-propan-2-yl-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-yl]-2-methoxy-phenyl]-~{N}-(2-azanyl-2-oxidanylidene-ethyl)benzamide, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Singh, A.K, Anthonyrajah, E.S, Brown, D.G. | | Deposit date: | 2016-06-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Targeting a Subpocket in Trypanosoma brucei Phosphodiesterase B1 (TbrPDEB1) Enables the Structure-Based Discovery of Selective Inhibitors with Trypanocidal Activity.
J. Med. Chem., 61, 2018
|
|
5L8G
 
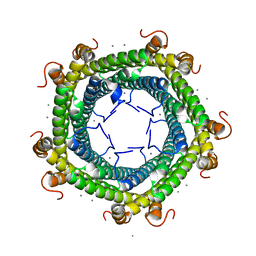 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant H65A | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | He, D, Hughes, S, Vanden-Hehir, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2016-06-07 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.974 Å) | | Cite: | Structural characterization of encapsulated ferritin provides insight into iron storage in bacterial nanocompartments.
Elife, 5, 2016
|
|
4YWC
 
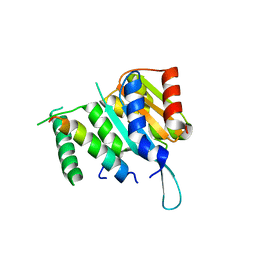 | | Crystal structure of Myc3(5-242) fragment in complex with Jaz9(218-239) peptide | | Descriptor: | Protein TIFY 7, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Brunzelle, J.S, Xu, H.E, Melcher, K, He, S.Y. | | Deposit date: | 2015-03-20 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
8FOG
 
 | |
5L96
 
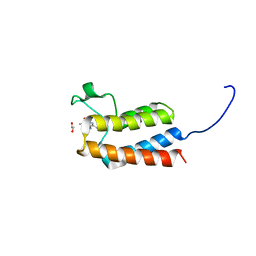 | | Crystal Structure of BAZ2B bromodomain in complex with 3-amino-2-methylpyridine derivative 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-~{N}-[(2~{R})-1-methylsulfonylpropan-2-yl]pyridin-3-amine, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Lolli, G, Marchand, J.-R, Caflisch, A. | | Deposit date: | 2016-06-09 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Derivatives of 3-Amino-2-methylpyridine as BAZ2B Bromodomain Ligands: In Silico Discovery and in Crystallo Validation.
J. Med. Chem., 59, 2016
|
|
6VWF
 
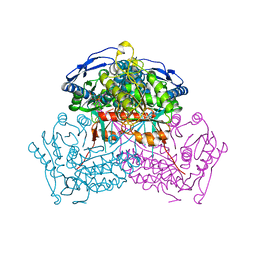 | | Structure of ALDH9A1 complexed with NAD+ in space group C222 | | Descriptor: | 4-trimethylaminobutyraldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Wyatt, J.W, Tanner, J.J. | | Deposit date: | 2020-02-19 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Inhibition, crystal structures, and in-solution oligomeric structure of aldehyde dehydrogenase 9A1.
Arch.Biochem.Biophys., 691, 2020
|
|
5LA1
 
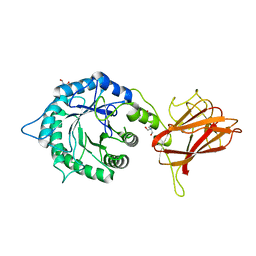 | | The mechanism by which arabinoxylanases can recognise highly decorated xylans | | Descriptor: | CALCIUM ION, Carbohydrate binding family 6, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM, ... | | Authors: | Basle, A, Labourel, A, Cuskin, F, Jackson, A, Crouch, L, Rogowski, A, Gilbert, H. | | Deposit date: | 2016-06-13 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Mechanism by Which Arabinoxylanases Can Recognize Highly Decorated Xylans.
J.Biol.Chem., 291, 2016
|
|
8FNY
 
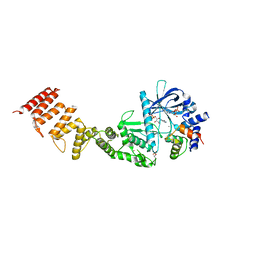 | | Nucleotide-bound structure of a functional construct of eukaryotic elongation factor 2 kinase. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Piserchio, A, Isiorho, E.A, Dalby, K.N, Ghose, R. | | Deposit date: | 2022-12-28 | | Release date: | 2023-05-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | ADP enhances the allosteric activation of eukaryotic elongation factor 2 kinase by calmodulin.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5LAE
 
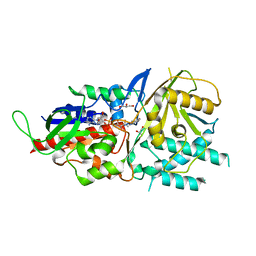 | | Crystal structure of murine N1-acetylpolyamine oxidase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Peroxisomal N(1)-acetyl-spermine/spermidine oxidase,Peroxisomal N(1)-acetyl-spermine/spermidine oxidase | | Authors: | Sjogren, T, Aagaard, A, Snijder, A, Barlind, L. | | Deposit date: | 2016-06-14 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of Murine N(1)-Acetylspermine Oxidase Reveals Molecular Details of Vertebrate Polyamine Catabolism.
Biochemistry, 56, 2017
|
|
5LAQ
 
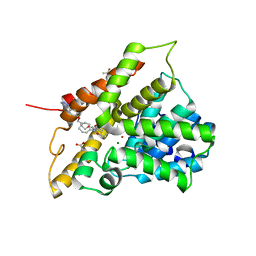 | | Crystal structure of human phosphodiesterase 4B catalytic domain with inhibitor NPD-001 | | Descriptor: | (4~{a}~{S},8~{a}~{R})-2-cycloheptyl-4-[4-methoxy-3-[4-[4-(1~{H}-1,2,3,4-tetrazol-5-yl)phenoxy]butoxy]phenyl]-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-one, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2016-06-14 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting a Subpocket in Trypanosoma brucei Phosphodiesterase B1 (TbrPDEB1) Enables the Structure-Based Discovery of Selective Inhibitors with Trypanocidal Activity.
J. Med. Chem., 61, 2018
|
|
8FJ9
 
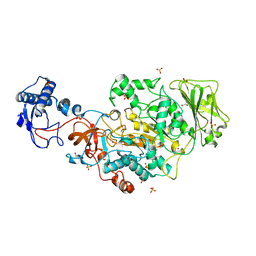 | | Structure of the catalytic domain of Streptococcus mutans GtfB in tetragonal space group P4322 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schormann, N, Deivanayagam, C. | | Deposit date: | 2022-12-19 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The catalytic domains of Streptococcus mutans glucosyltransferases: a structural analysis.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8FJC
 
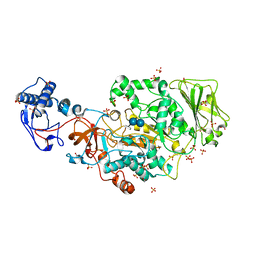 | | Structure of the catalytic domain of Streptococcus mutans GtfB complexed to acarbose in tetragonal space group P4322 | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, ... | | Authors: | Schormann, N, Deivanayagam, C. | | Deposit date: | 2022-12-19 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The catalytic domains of Streptococcus mutans glucosyltransferases: a structural analysis.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
5LBP
 
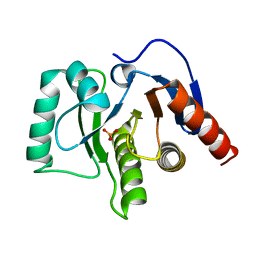 | | Oceanobacillus iheyensis macrodomain mutant N30A | | Descriptor: | MacroD-type macrodomain, PHOSPHATE ION | | Authors: | Gil-Ortiz, F, Zapata-Perez, R, Martinez, A.B, Juanhuix, J, Sanchez-Ferrer, A. | | Deposit date: | 2016-06-16 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and functional analysis of Oceanobacillus iheyensis macrodomain reveals a network of waters involved in substrate binding and catalysis.
Open Biol, 7, 2017
|
|
6W0A
 
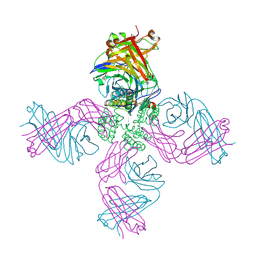 | | Open-gate KcsA soaked in 1 mM BaCl2 | | Descriptor: | BARIUM ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.237 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
6W0I
 
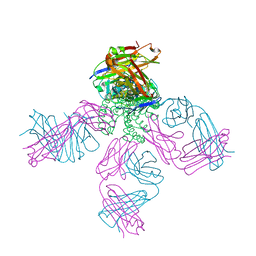 | | Closed-gate KcsA soaked in 10mM KCl/5mM BaCl2 | | Descriptor: | Fab Heavy Chain, Fab Light Chain, POTASSIUM ION, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.328 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
8FK4
 
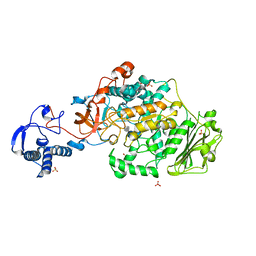 | | Structure of the catalytic domain of Streptococcus mutans GtfB complexed to acarbose in orthorhombic space group P21212 | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, Glucosyltransferase-I, ... | | Authors: | Schormann, N, Deivanayagam, C. | | Deposit date: | 2022-12-20 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The catalytic domains of Streptococcus mutans glucosyltransferases: a structural analysis.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
