4PV7
 
 | | Cocrystal structure of dipeptidyl-peptidase 4 with an indole scaffold inhibitor | | Descriptor: | 1-[2-(2,4-dichlorophenyl)-1-(methylsulfonyl)-1H-indol-3-yl]methanamine, Dipeptidyl peptidase 4 soluble form | | Authors: | Xiao, P, Guo, R, Huang, S, Cui, H, Ye, S, Zhang, Z. | | Deposit date: | 2014-03-15 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Discovery of dipeptidyl peptidase IV (DPP4) inhibitors based on a novel indole scaffold
Chin.Chem.Lett., 25, 2014
|
|
4P9T
 
 | | Structure of the free form of the N-terminal VH1 domain of monomeric alpha-catenin | | Descriptor: | 1,2-ETHANEDIOL, Catenin alpha-2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shibahara, T, Hirano, Y, Hakoshima, T. | | Deposit date: | 2014-04-04 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the free form of the N-terminal VH1 domain of monomeric alpha-catenin.
Febs Lett., 589, 2015
|
|
4MX3
 
 | | Crystal Structure of PKA RIalpha Homodimer | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Bruystens, J.G.H, Wu, J, Fortezzo, A, Kornev, A.P, Blumenthal, D.A, Taylor, S.S. | | Deposit date: | 2013-09-25 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.88 Å) | | Cite: | PKA RI alpha Homodimer Structure Reveals an Intermolecular Interface with Implications for Cooperative cAMP Binding and Carney Complex Disease.
Structure, 22, 2014
|
|
4MYR
 
 | |
4MU9
 
 | |
4MRU
 
 | |
4N9C
 
 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 5-nitro-1H-benzimidazole, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhao, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | Deposit date: | 2013-10-20 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4MUD
 
 | |
4MJG
 
 | |
4N0M
 
 | | Crystal structure of human C53A DJ-1 in complex with Cu | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Protein DJ-1 | | Authors: | Cendron, L, Girotto, S, Bisaglia, M, Tessari, I, Mammi, S, Zanotti, G, Bubacco, L. | | Deposit date: | 2013-10-02 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | DJ-1 Is a Copper Chaperone Acting on SOD1 Activation.
J.Biol.Chem., 289, 2014
|
|
4N0P
 
 | |
4OBM
 
 | |
4P34
 
 | | Crystal structure of DJ-1 in sulfenic acid form (fresh crystal) | | Descriptor: | PENTAETHYLENE GLYCOL, Protein DJ-1 | | Authors: | Tashiro, S, Wu, C.-X, Hoang, Q.Q, Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Thermodynamic and Structural Characterization of the Specific Binding of Zn(II) to Human Protein DJ-1.
Biochemistry, 53, 2014
|
|
4P4B
 
 | |
4P4F
 
 | |
4PXF
 
 | |
4Q08
 
 | | Crystal structure of chimeric carbonic anhydrase XII with inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-(cyclooctylamino)-3,5,6-trifluoro-4-[(2-hydroxyethyl)sulfanyl]benzenesulfonamide, BICINE, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-04-01 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Discovery and characterization of novel selective inhibitors of carbonic anhydrase IX.
J.Med.Chem., 57, 2014
|
|
4PXT
 
 | |
4Q5O
 
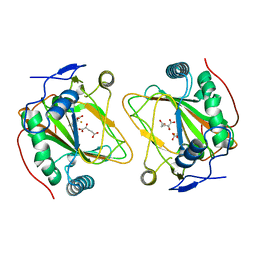 | | Crystal structure of EctD from S. alaskensis with 2-oxoglutarate and 5-hydroxyectoine | | Descriptor: | (4S,5S)-5-HYDROXY-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, 2-OXOGLUTARIC ACID, Ectoine hydroxylase, ... | | Authors: | Hoeppner, A, Widderich, N, Bremer, E, Smits, S.H. | | Deposit date: | 2014-04-17 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of the ectoine hydroxylase, a snapshot of the active site.
J.Biol.Chem., 289, 2014
|
|
4Q0L
 
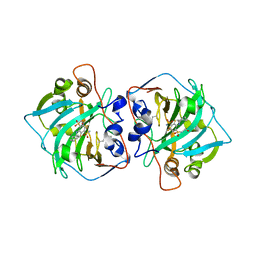 | | Crystal structure of catalytic domain of human carbonic anhydrase isozyme XII with inhibitor | | Descriptor: | 3-(cyclooctylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 12, ZINC ION | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-04-02 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and characterization of novel selective inhibitors of carbonic anhydrase IX.
J.Med.Chem., 57, 2014
|
|
4Q0Y
 
 | |
4QNW
 
 | |
4Q6K
 
 | |
4PZH
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 2,3,5,6-tetrafluoro-4[(2-hydroxyethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 2,3,5,6-tetrafluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-03-31 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Discovery and characterization of novel selective inhibitors of carbonic anhydrase IX.
J.Med.Chem., 57, 2014
|
|
4Q9H
 
 | | P-glycoprotein at 3.4 A resolution | | Descriptor: | Multidrug resistance protein 1A | | Authors: | McGrath, A.P, Szewczyk, P, Chang, G. | | Deposit date: | 2014-05-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
