1OWO
 
 | | DATA4:photoreduced DNA photolyase / received X-rays dose 1.2 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1P1D
 
 | |
1O8Y
 
 | | Solution structure of SFTI-1(6,5), an acyclic permutant of the proteinase inhibitor SFTI-1, trans-trans-trans conformer (tt-A) | | Descriptor: | CYCLIC TRYPSIN INHIBITOR | | Authors: | Marx, U.C, Craik, D.J. | | Deposit date: | 2002-12-09 | | Release date: | 2003-03-13 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Enzymatic Cyclization of a Potent Bowman-Birk Protease Inhibitor, Sunflower Trypsin Inhibitor-1, and Solution Structure of an Acyclic Precursor Peptide
J.Biol.Chem., 278, 2003
|
|
1OW6
 
 | | Paxillin LD4 motif bound to the Focal Adhesion Targeting (FAT) domain of the Focal Adhesion Kinase | | Descriptor: | Focal adhesion kinase 1, Paxillin | | Authors: | Hoellerer, M.K, Noble, M.E.M, Labesse, G, Campbell, I.D, Werner, J.M, Arold, S.T. | | Deposit date: | 2003-03-28 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular Recognition of Paxillin LD motifs
by the Focal Adhesion Targeting Domain
Structure, 11, 2003
|
|
1OWT
 
 | | Structure of the Alzheimer's disease amyloid precursor protein copper binding domain | | Descriptor: | Amyloid beta A4 protein | | Authors: | Barnham, K.J, McKinstry, W.J, Multhaup, G, Galatis, D, Morton, C.J, Curtain, C.C, Williamson, N.A, White, A.R, Hinds, M.G, Norton, R.S, Beyreuther, K, Masters, C.L, Parker, M.W, Cappai, R. | | Deposit date: | 2003-03-30 | | Release date: | 2003-05-13 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure of the Alzheimer's Disease Amyloid Precursor Protein Copper Binding Domain. A REGULATOR OF NEURONAL COPPER HOMEOSTASIS.
J.Biol.Chem., 278, 2003
|
|
1OPI
 
 | | SOLUTION STRUCTURE OF THE THIRD RNA RECOGNITION MOTIF (RRM) OF U2AF65 IN COMPLEX WITH AN N-TERMINAL SF1 PEPTIDE | | Descriptor: | SPLICING FACTOR SF1, SPLICING FACTOR U2AF 65 KDA SUBUNIT | | Authors: | Selenko, P, Gregorovic, G, Sprangers, R, Stier, G, Rhani, Z, Kramer, A, Sattler, M. | | Deposit date: | 2003-03-05 | | Release date: | 2004-03-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP
Mol.Cell, 11, 2003
|
|
1P88
 
 | |
1NWV
 
 | | SOLUTION STRUCTURE OF A FUNCTIONALLY ACTIVE COMPONENT OF DECAY ACCELERATING FACTOR | | Descriptor: | Complement decay-accelerating factor | | Authors: | Uhrinova, S, Lin, F, Ball, G, Bromek, K, Uhrin, D, Medof, M.E, Barlow, P.N. | | Deposit date: | 2003-02-07 | | Release date: | 2003-04-22 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a functionally active fragment of decay-accelerating factor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1M39
 
 | | Solution structure of the C-terminal fragment (F86-I165) of the human centrin 2 in calcium saturated form | | Descriptor: | Caltractin, isoform 1 | | Authors: | Matei, E, Miron, S, Blouquit, Y, Duchambon, P, Durussel, P, Cox, J.A, Craescu, C.T. | | Deposit date: | 2002-06-27 | | Release date: | 2003-03-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | C-terminal half of human centrin 2 behaves like a regulatory EF-hand domain
Biochemistry, 42, 2003
|
|
1M9L
 
 | | Relaxation-based Refined Structure Of Chlamydomonas Outer Arm Dynein Light Chain 1 | | Descriptor: | Outer Arm Dynein Light Chain 1 | | Authors: | Wu, H.W, Maciejewski, M.W, Marintchev, A, Benashski, S.E, Mullen, G.P, King, S.M. | | Deposit date: | 2002-07-29 | | Release date: | 2003-03-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Relaxation-based structure refinement and backbone molecular dynamics of the
Dynein motor domain-associated light chain
Biochemistry, 42, 2003
|
|
1MHU
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF HUMAN [113CD7] METALLOTHIONEIN-2 IN SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2 | | Authors: | Braun, W, Messerle, B.A, Schaeffer, A, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of human [113Cd7]metallothionein-2 in solution determined by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 214, 1990
|
|
1P1E
 
 | |
1OW8
 
 | | Paxillin LD2 motif bound to the Focal Adhesion Targeting (FAT) domain of the Focal Adhesion Kinase | | Descriptor: | Focal adhesion kinase 1, Paxillin | | Authors: | Hoellerer, M.K, Noble, M.E.M, Labesse, G, Werner, J.M, Arold, S.T. | | Deposit date: | 2003-03-28 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular Recognition of Paxillin LD Motifs
by the Focal Adhesion Targeting Domain
Structure, 11, 2003
|
|
1OWP
 
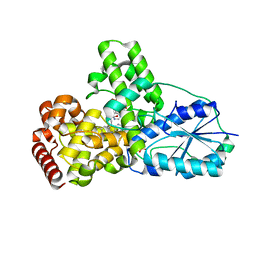 | | DATA6:photoreduced DNA pholyase / received X-rays dose 4.8 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OWL
 
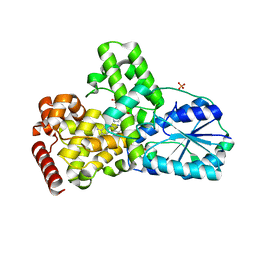 | | Structure of apophotolyase from Anacystis nidulans | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PKS
 
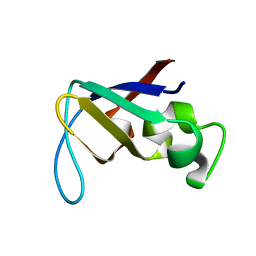 | | STRUCTURE OF THE PI3K SH3 DOMAIN AND ANALYSIS OF THE SH3 FAMILY | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT SH3 DOMAIN | | Authors: | Koyama, S, Yu, H, Dalgarno, D.C, Shin, T.B, Zydowsky, L.D, Schreiber, S.L. | | Deposit date: | 1994-03-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the PI3K SH3 domain and analysis of the SH3 family.
Cell(Cambridge,Mass.), 72, 1993
|
|
1MFA
 
 | |
1OW7
 
 | | Paxillin LD4 motif bound to the Focal Adhesion Targeting (FAT) domain of the Focal Adhesion Kinase | | Descriptor: | Focal adhesion kinase 1, Paxillin | | Authors: | Hoellerer, M.K, Noble, M.E.M, Labesse, G, Werner, J.M, Arold, S.T. | | Deposit date: | 2003-03-28 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Recognition of Paxillin LD Motifs
by the Focal Adhesion Targeting Domain
Structure, 11, 2003
|
|
1OWM
 
 | | DATA1:DNA photolyase / received X-rays dose 1.2 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1Q68
 
 | | Solution structure of T-cell surface glycoprotein CD4 and Proto-oncogene tyrosine-protein kinase LCK fragments | | Descriptor: | Proto-oncogene tyrosine-protein kinase LCK, T-cell surface glycoprotein CD4, ZINC ION | | Authors: | Kim, P.W, Sun, Z.Y, Blacklow, S.C, Wagner, G, Eck, M.J. | | Deposit date: | 2003-08-12 | | Release date: | 2003-11-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A zinc clasp structure tethers Lck to T cell coreceptors CD4 and CD8.
Science, 301, 2003
|
|
1PKT
 
 | | STRUCTURE OF THE PI3K SH3 DOMAIN AND ANALYSIS OF THE SH3 FAMILY | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT SH3 DOMAIN | | Authors: | Koyama, S, Yu, H, Dalgarno, D.C, Shin, T.B, Zydowsky, L.D, Schreiber, S.L. | | Deposit date: | 1994-03-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the PI3K SH3 domain and analysis of the SH3 family.
Cell(Cambridge,Mass.), 72, 1993
|
|
1PNN
 
 | | PEPTIDE NUCLEIC ACID (PNA) COMPLEXED WITH DNA | | Descriptor: | DNA (5'-D(GP*AP*AP*GP*AP*AP*GP*AP*G)-3'), PNA (NH2-P(*C*T*C*T*T*C*T*T*C-HIS-GLY-SER-SER-GLY-HIS-C*T*T*C*T*T*C*T*C)-COOH) | | Authors: | Betts, L, Veal, J.M. | | Deposit date: | 1995-10-13 | | Release date: | 1996-03-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Nucleic Acid Triple Helix Formed by a Peptide Nucleic Acid-DNA Complex
Science, 270, 1995
|
|
1PUP
 
 | |
1MEA
 
 | |
1MED
 
 | |
