8BYO
 
 | | fragment-linked stabilizer for ERa - 14-3-3 interaction (1074361) | | Descriptor: | 14-3-3 protein sigma, ERalpha peptide, MAGNESIUM ION, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2023-01-18 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6ZUQ
 
 | | Crystal structure of the effector Ecp11-1 from Fulvia fulva | | Descriptor: | Extracellular protein 11-1, GLYCEROL, ZINC ION | | Authors: | Lazar, N, Mesarich, C, Petit-Houdenot, Y, Talbi, N, Li de la Sierra-Gallay, I, Zelie, E, Blondeau, K, Gracy, J, Ollivier, B, van de Wouw, A, Balesdent, M.H, Idnurm, A, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2020-07-23 | | Release date: | 2021-08-04 | | Last modified: | 2022-07-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins.
Plos Pathog., 18, 2022
|
|
7R0K
 
 | | Crystal structure of Polymerase I from phage G20c | | Descriptor: | DNA polymerase I | | Authors: | Welin, M, Svensson, A, Hakansson, M, Al-Karadaghi, S, Linares-Pasten, J.A, Jasilionis, A, Nordberg Karlsson, E, Ahlqvist, J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.972 Å) | | Cite: | Crystal structure of DNA polymerase I from Thermus phage G20c.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8BZW
 
 | | Co-soaked stabilizers for ERa - 14-3-3 interaction (844_AZ210) | | Descriptor: | 14-3-3 protein sigma, 2-(4-chloranylphenoxy)-2-methyl-~{N}-(2-sulfanylethyl)propanamide, 4-[(2~{R})-3-azanyl-2-methyl-propyl]-7-methoxy-1-benzothiophene-2-carboximidamide, ... | | Authors: | Visser, E.J, Sijbesma, E, Ottmann, C. | | Deposit date: | 2022-12-15 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8K6J
 
 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus variant-H1147A | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase (H1147A) large subunit, ... | | Authors: | Fukawa, E, Miyata, T, Makino, F, Adachi, T, Suzuki, Y, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2023-07-25 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural and electrochemical elucidation of biocatalytic mechanisms in direct electron transfer-type D-fructose dehydrogenase.
Electrochim Acta, 490, 2024
|
|
6ZUS
 
 | | Crystal structure of the effector Ecp11-1 from Fulvia fulva | | Descriptor: | DI(HYDROXYETHYL)ETHER, Extracellular protein 11-1, GLYCEROL, ... | | Authors: | Lazar, N, Mesarich, C, Petit-Houdenot, Y, Talbi, N, Li de la Sierra-Gallay, I, Zelie, E, Blondeau, K, Gracy, J, Ollivier, B, van de Wouw, A, Balesdent, M.H, Idnurm, A, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2020-07-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins.
Plos Pathog., 18, 2022
|
|
6Y6K
 
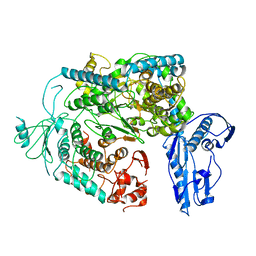 | | Cryo-EM structure of a Phenuiviridae L protein | | Descriptor: | MAGNESIUM ION, RNA-dependent RNA polymerase | | Authors: | Vogel, D, Thorkelsson, S.R, Quemin, E, Meier, K, Kouba, T, Gogrefe, N, Busch, C, Reindl, S, Guenther, S, Cusack, S, Gruenewald, K, Rosenthal, M. | | Deposit date: | 2020-02-26 | | Release date: | 2020-04-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structural and functional characterization of the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 48, 2020
|
|
5KAF
 
 | | RT XFEL structure of Photosystem II in the dark state at 3.0 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.00001 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
8PKJ
 
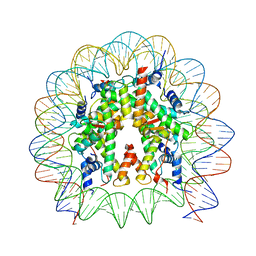 | | Cryo-EM structure of the nucleosome containing Nr5a2 motif at SHL+5.5 | | Descriptor: | DNA, Histone H2A, Histone H2B, ... | | Authors: | Kobayashi, W, Sappler, A, Bollschweiler, D, Kummecke, M, Basquin, J, Arslantas, E, Ruangroengkulrith, S, Hornberger, R, Duderstadt, K, Tachibana, K. | | Deposit date: | 2023-06-26 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Nucleosome-bound NR5A2 structure reveals pioneer factor mechanism by DNA minor groove anchor competition.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8K6K
 
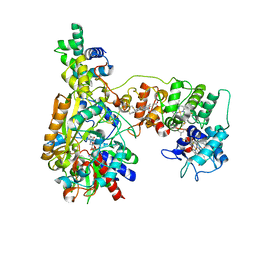 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus variant-N1146A | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase (N1146A) large subunit, ... | | Authors: | Fukawa, E, Miyata, T, Makino, F, Adachi, T, Suzuki, Y, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2023-07-25 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural and electrochemical elucidation of biocatalytic mechanisms in direct electron transfer-type D-fructose dehydrogenase.
Electrochim Acta, 490, 2024
|
|
8BYE
 
 | |
7R67
 
 | | Human obscurin Ig13 | | Descriptor: | Obscurin | | Authors: | Moncure, G.E, Wright, N.T. | | Deposit date: | 2021-06-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The N-terminus of obscurin is flexible in solution.
Proteins, 91, 2023
|
|
7R68
 
 | | Human obscurin Ig12 | | Descriptor: | Obscurin | | Authors: | Mauriello, G.E, Wright, N.T. | | Deposit date: | 2021-06-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The N-terminus of obscurin is flexible in solution.
Proteins, 91, 2023
|
|
8BYB
 
 | |
8BX4
 
 | | fragment-linked stabilizer for ERa - 14-3-3 interaction (1074392) | | Descriptor: | 14-3-3 protein sigma, 2-[3,4-bis(chloranyl)phenoxy]-~{N}-[3-(5-carbamimidoylthiophen-3-yl)phenyl]-2-methyl-propanamide, ERalpha peptide | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-12-07 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8BXO
 
 | | fragment-linked stabilizer for ERa - 14-3-3 interaction (1075287) | | Descriptor: | 14-3-3 protein sigma, ERalpha peptide, ~{N}-[3-(5-carbamimidoylthiophen-3-yl)phenyl]-4,4-bis(fluoranyl)-1-phenoxy-cyclohexane-1-carboxamide | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-12-09 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8BXQ
 
 | |
8BY9
 
 | | fragment-linked stabilizer for ERa - 14-3-3 interaction (1075292) | | Descriptor: | 14-3-3 protein sigma, ERalpha peptide, ~{N}-[3-(5-carbamimidoylthiophen-3-yl)phenyl]-1-(4-chloranylphenoxy)-4,4-bis(fluoranyl)cyclohexane-1-carboxamide | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-12-12 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5K4Y
 
 | | Three-dimensional structure of L-threonine 3-dehydrogenase from Trypanosoma brucei refined to 1.77 angstroms | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Adjogatse, E.A, Erskine, P.T, Cooper, J.B. | | Deposit date: | 2016-05-22 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure and function of L-threonine-3-dehydrogenase from the parasitic protozoan Trypanosoma brucei revealed by X-ray crystallography and geometric simulations.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
8B9N
 
 | |
6ZZ4
 
 | | Crystal structure of the PTPN2 C216G mutant | | Descriptor: | PHOSPHATE ION, Tyrosine-protein phosphatase non-receptor type 2 | | Authors: | Mechaly, A.E, Berthelet, J, Nian, Q, Parlato, M, Cerf-Bensussan, N, Haouz, A, Rodrigues-Lima, F. | | Deposit date: | 2020-08-03 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural characterization of a pathogenic mutant of human protein tyrosine phosphatase PTPN2 (Cys216Gly) that causes very early onset autoimmune enteropathy.
Protein Sci., 31, 2022
|
|
8BXS
 
 | |
8BZB
 
 | | co-soak stabilizers for ERa - 14-3-3 interaction (884_AZ275) | | Descriptor: | 14-3-3 protein sigma, 2-(4-chloranylphenoxy)-2-methyl-~{N}-(2-sulfanylethyl)propanamide, 4-ethoxy-1-benzothiophene-2-carboximidamide, ... | | Authors: | Visser, E.J, Sijbesma, E, Ottmann, C. | | Deposit date: | 2022-12-14 | | Release date: | 2023-08-02 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6NRW
 
 | | Crystal structure of Dpr1 IG1 bound to DIP-eta IG1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cheng, S, Park, Y.J, Kurleto, J.D, Ozkan, E. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis of synaptic specificity by immunoglobulin superfamily receptors in Drosophila.
Elife, 8, 2019
|
|
6ZZK
 
 | |
