1EBF
 
 | | HOMOSERINE DEHYDROGENASE FROM S. CEREVISIAE COMPLEX WITH NAD+ | | Descriptor: | HOMOSERINE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | DeLaBarre, B, Thompson, P.R, Wright, G.D, Berghuis, A.M. | | Deposit date: | 2000-01-24 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of homoserine dehydrogenase suggest a novel catalytic mechanism for oxidoreductases.
Nat.Struct.Biol., 7, 2000
|
|
8CK4
 
 | | STRUCTURE OF HIF2A-ARNT HETERODIMER IN COMPLEX WITH (4S)-1-(3,5-difluorophenyl)-5,5-difluoro-3-methanesulfonyl-4,5,6,7-tetrahydro-2-benzothiophen-4-ol | | Descriptor: | (4~{S})-1-[3,5-bis(fluoranyl)phenyl]-5,5-bis(fluoranyl)-3-methylsulfonyl-6,7-dihydro-4~{H}-2-benzothiophen-4-ol, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Musil, D. | | Deposit date: | 2023-02-14 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of Cycloalkyl[ c ]thiophenes as Novel Scaffolds for Hypoxia-Inducible Factor-2 alpha Inhibitors.
J.Med.Chem., 66, 2023
|
|
7R0J
 
 | | Structure of the V2 receptor Cter-arrestin2-ScFv30 complex | | Descriptor: | Arrestin2, ScFv30, V2R Cter | | Authors: | Bous, J, Fouillen, A, Trapani, S, Granier, S, Mouillac, B, Bron, P. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structure of the vasopressin hormone-V2 receptor-beta-arrestin1 ternary complex.
Sci Adv, 8, 2022
|
|
7R0C
 
 | | Structure of the AVP-V2R-arrestin2-ScFv30 complex | | Descriptor: | AVP, Arrestin2, ScFv30, ... | | Authors: | Bous, J, Fouillen, A, Trapani, S, Granier, S, Mouillac, B, Bron, P. | | Deposit date: | 2022-02-01 | | Release date: | 2022-09-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.73 Å) | | Cite: | Structure of the vasopressin hormone-V2 receptor-beta-arrestin1 ternary complex.
Sci Adv, 8, 2022
|
|
3O84
 
 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 6-phenyl-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 6-phenyl-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid, ... | | Authors: | Drake, E.J, Duckworth, B.P, Neres, J, Aldrich, C.C, Gulick, A.M. | | Deposit date: | 2010-08-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and structural characterization of bisubstrate inhibitors of BasE, the self-standing nonribosomal peptide synthetase adenylate-forming enzyme of acinetobactin synthesis.
Biochemistry, 49, 2010
|
|
4Y5Y
 
 | | Diabody 330 complex with EpoR | | Descriptor: | Erythropoietin receptor, GLYCEROL, diabody 330 VH domain, ... | | Authors: | Moraga, I, Guo, F, Ozkan, E, Jude, K.M, Garcia, K.C. | | Deposit date: | 2015-02-12 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Tuning Cytokine Receptor Signaling by Re-orienting Dimer Geometry with Surrogate Ligands.
Cell, 160, 2015
|
|
4JJB
 
 | |
1FB5
 
 | | LOW RESOLUTION STRUCTURE OF OVINE ORNITHINE TRANSCARBMOYLASE IN THE UNLIGANDED STATE | | Descriptor: | NORVALINE, ORNITHINE TRANSCARBAMOYLASE | | Authors: | Zanotti, G, Battistutta, R, Panzalorto, M, Francescato, P, Bruno, G, De Gregorio, A. | | Deposit date: | 2000-07-14 | | Release date: | 2003-08-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Functional and structural characterization of ovine ornithine transcarbamoylase.
Org.Biomol.Chem., 1, 2003
|
|
1BMP
 
 | | BONE MORPHOGENETIC PROTEIN-7 | | Descriptor: | BONE MORPHOGENETIC PROTEIN-7 | | Authors: | Griffith, D.L, Scott, D.L. | | Deposit date: | 1995-12-14 | | Release date: | 1997-07-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of recombinant human osteogenic protein 1: structural paradigm for the transforming growth factor beta superfamily.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1UP5
 
 | | Chicken Calmodulin | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Wilson, M.A, Rupp, B. | | Deposit date: | 2003-09-27 | | Release date: | 2005-03-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallization and Preliminary X-Ray Analysis of Two New Crystal Forms of Calmodulin
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1CGL
 
 | | Structure of the catalytic domain of fibroblast collagenase complexed with an inhibitor | | Descriptor: | CALCIUM ION, FIBROBLAST COLLAGENASE, N-[(1S)-3-{[(benzyloxy)carbonyl]amino}-1-carboxypropyl]-L-leucyl-N-(2-morpholin-4-ylethyl)-L-phenylalaninamide, ... | | Authors: | Lovejoy, B, Cleasby, A, Hassell, A.M, Longley, K, Luther, M.A, Weigl, D, Mcgeehan, G, Mcelroy, A.B, Drewry, D, Lambert, M.H, Jordan, S.R. | | Deposit date: | 1993-11-17 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the catalytic domain of fibroblast collagenase complexed with an inhibitor.
Science, 263, 1994
|
|
3P6I
 
 | |
7QTZ
 
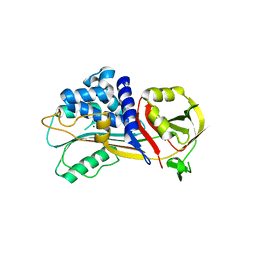 | | Crystal structure of Iripin-1 serpin from tick Ixodes ricinus | | Descriptor: | MAGNESIUM ION, Putative salivary serpin | | Authors: | Kascakova, B, Kuta Smatanova, I, Chmelar, J, Prudnikova, T. | | Deposit date: | 2022-01-17 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Iripin-1, a new anti-inflammatory tick serpin, inhibits leukocyte recruitment in vivo while altering the levels of chemokines and adhesion molecules.
Front Immunol, 14, 2023
|
|
8R4A
 
 | |
3ALO
 
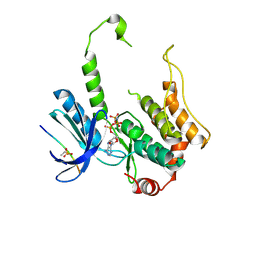 | | Crystal structure of human non-phosphorylated MKK4 kinase domain ternary complex with AMP-PNP and p38 peptide | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 4, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Matsumoto, T, Kinoshita, T, Kirii, Y, Yokota, K, Hamada, K, Tada, T. | | Deposit date: | 2010-08-04 | | Release date: | 2010-10-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of MKK4 kinase domain reveal that substrate peptide binds to an allosteric site and induces an auto-inhibition state
Biochem.Biophys.Res.Commun., 400, 2010
|
|
1CLX
 
 | | CATALYTIC CORE OF XYLANASE A | | Descriptor: | CALCIUM ION, XYLANASE A | | Authors: | Harris, G.W, Jenkins, J.A, Connerton, I, Pickersgill, R.W. | | Deposit date: | 1995-08-31 | | Release date: | 1996-06-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined crystal structure of the catalytic domain of xylanase A from Pseudomonas fluorescens at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
7ZLJ
 
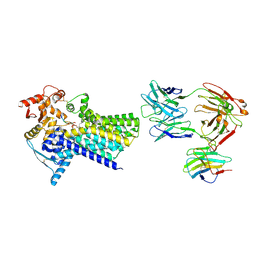 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in ternary complex with Dol25-P-C-Man and acceptor peptide, bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mao, R, Mukherjee, S, Boilevin, J, Irobalieva, R, Darbre, T, Reymond, J.L, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
2YMA
 
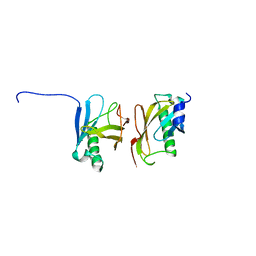 | | X-ray structure of the Yos9 dimerization domain | | Descriptor: | PROTEIN OS-9 HOMOLOG | | Authors: | Hanna, J, Schuetz, A, Zimmermann, F, Behlke, J, Sommer, T, Heinemann, U. | | Deposit date: | 2011-06-07 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | Structural and Biochemical Basis of Yos9 Protein Dimerization and Possible Contribution to Self-Association of 3-Hydroxy-3-Methylglutaryl-Coenzyme a Reductase Degradation Ubiquitin-Ligase Complex.
J.Biol.Chem., 287, 2012
|
|
5NWX
 
 | | Insight into the molecular recognition mechanism of the coactivator NCoA1 by STAT6 | | Descriptor: | Nuclear receptor coactivator 1, Signal transducer and activator of transcription 6 | | Authors: | Russo, L, Giller, K, Pfitzner, E, Griesinger, C, Becker, S. | | Deposit date: | 2017-05-08 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Insight into the molecular recognition mechanism of the coactivator NCoA1 by STAT6.
Sci Rep, 7, 2017
|
|
8UDL
 
 | | Human Mitochondrial DNA Polymerase Gamma Binary Complex | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*CP*GP*AP*CP*GP*GP*CP*CP*AP*GP*TP*GP*CP*CP*AP*TP*AP*C)-3'), DNA (5'-D(P*AP*GP*GP*TP*AP*TP*GP*GP*CP*AP*CP*TP*GP*GP*CP*CP*GP*TP*CP*GP*TP*TP*TP*T)-3'), DNA polymerase subunit gamma-1, ... | | Authors: | Park, J, Yin, Y.W. | | Deposit date: | 2023-09-28 | | Release date: | 2024-06-05 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | An interaction network in the polymerase active site is a prerequisite for Watson-Crick base pairing in Pol gamma.
Sci Adv, 10, 2024
|
|
3ICB
 
 | | THE REFINED STRUCTURE OF VITAMIN D-DEPENDENT CALCIUM-BINDING PROTEIN FROM BOVINE INTESTINE. MOLECULAR DETAILS, ION BINDING, AND IMPLICATIONS FOR THE STRUCTURE OF OTHER CALCIUM-BINDING PROTEINS | | Descriptor: | CALCIUM ION, CALCIUM-BINDING PROTEIN, SULFATE ION | | Authors: | Szebenyi, D.M.E, Moffat, K. | | Deposit date: | 1986-09-09 | | Release date: | 1986-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The refined structure of vitamin D-dependent calcium-binding protein from bovine intestine. Molecular details, ion binding, and implications for the structure of other calcium-binding proteins.
J.Biol.Chem., 261, 1986
|
|
1ITM
 
 | | ANALYSIS OF THE SOLUTION STRUCTURE OF HUMAN INTERLEUKIN 4 DETERMINED BY HETERONUCLEAR THREE-DIMENSIONAL NUCLEAR MAGNETIC RESONANCE TECHNIQUES | | Descriptor: | INTERLEUKIN-4 | | Authors: | Redfield, C, Smith, L.J, Boyd, J, Lawrence, G.M.P, Edwards, R.G, Gershater, C.J, Smith, R.A.G, Dobson, C.M. | | Deposit date: | 1994-02-28 | | Release date: | 1994-05-31 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Analysis of the solution structure of human interleukin-4 determined by heteronuclear three-dimensional nuclear magnetic resonance techniques.
J.Mol.Biol., 238, 1994
|
|
5NFT
 
 | | Glucocorticoid Receptor in complex with AZD5423 | | Descriptor: | 1,2-ETHANEDIOL, 2,2,2-tris(fluoranyl)-~{N}-[(1~{R},2~{S})-1-[1-(4-fluorophenyl)indazol-5-yl]oxy-1-(3-methoxyphenyl)propan-2-yl]ethanamide, Glucocorticoid receptor, ... | | Authors: | Edman, K, Wissler, L. | | Deposit date: | 2017-03-15 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Selective Nonsteroidal Glucocorticoid Receptor Modulators for the Inhaled Treatment of Pulmonary Diseases.
J. Med. Chem., 60, 2017
|
|
4RYT
 
 | |
2WDE
 
 | | Termus thermophilus Sulfate thiohydrolase SoxB in complex with thiosulfate | | Descriptor: | MANGANESE (II) ION, SULFUR OXIDATION PROTEIN SOXB, TERTIARY-BUTYL ALCOHOL, ... | | Authors: | Sauve, V, Roversi, P, Leath, K.J, Garman, E.F, Antrobus, R, Lea, S.M, Berks, B.C. | | Deposit date: | 2009-03-24 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism for the Hydrolysis of a Sulfur-Sulfur Bond Based on the Crystal Structure of the Thiosulfohydrolase Soxb.
J.Biol.Chem., 284, 2009
|
|
