8BW5
 
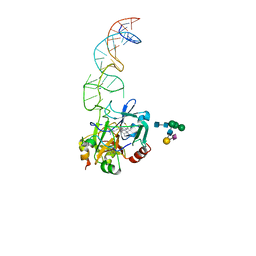 | | X-ray structure of the complex between human alpha thrombin and the duplex/quadruplex aptamer M08s-1_41mer | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, M08s-1_41mer, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Troisi, R, Napolitano, V, Sica, F. | | Deposit date: | 2022-12-06 | | Release date: | 2023-07-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Steric hindrance and structural flexibility shape the functional properties of a guanine-rich oligonucleotide.
Nucleic Acids Res., 51, 2023
|
|
6ZGO
 
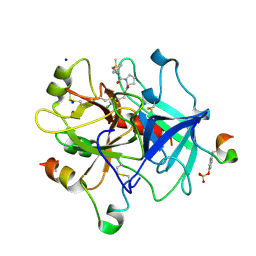 | | Thrombin in complex with D-Phe-Pro-2-chlorofuran derivative (13l) | | Descriptor: | (2~{S})-1-[(2~{R})-2-azanyl-3-phenyl-propanoyl]-~{N}-[(5-chloranylfuran-2-yl)methyl]pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Sandner, A, Heine, A, Klebe, G. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Thrombin in complex with D-Phe-Pro-2-chlorofuran derivative (13l)
To be published
|
|
6QHJ
 
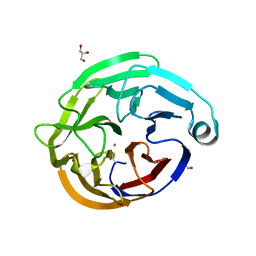 | | High-resolution crystal structure of calcium- and sodium-bound mouse Olfactomedin-1 beta-propeller domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Pronker, M.F, van den Hoek, H.G, Janssen, B.J.C. | | Deposit date: | 2019-01-16 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Design and structural characterisation of olfactomedin-1 variants as tools for functional studies.
BMC Mol Cell Biol, 20, 2019
|
|
4YS2
 
 | | RCK domain with CDA | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Na+/H+ antiporter-like protein | | Authors: | Chin, K.H, Chou, S.H. | | Deposit date: | 2015-03-16 | | Release date: | 2016-04-27 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | RCK domain with CDA
To Be Published
|
|
6CX9
 
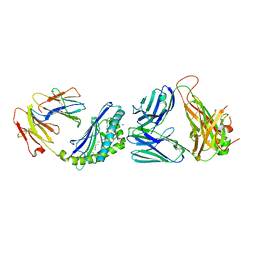 | | Structure of alpha-GSA[16,6P] bound by CD1d and in complex with the Va14Vb8.2 TCR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Wang, J, Zajonc, D. | | Deposit date: | 2018-04-02 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
6CX5
 
 | | Structure of alpha-GSA[8,8P] bound by CD1d and in complex with the Va14Vb8.2 TCR | | Descriptor: | (5R,6S,7S)-5,6-dihydroxy-7-(octanoylamino)-N-(8-phenyloctyl)-8-{[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]oxy}octanamide (non-preferred name), 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, J, Zajonc, D. | | Deposit date: | 2018-04-02 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
7ZO4
 
 | | L1 metallo-beta-lactamase in complex with hydrolysed panipenem | | Descriptor: | (2R,4S)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S)-1-ethanimidoylpyrrolidin-3-yl]sulfanyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Metallo-beta-lactamase L1, SODIUM ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-04-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Interactions of hydrolyzed beta-lactams with the L1 metallo-beta-lactamase: Crystallography supports stereoselective binding of cephem/carbapenem products.
J.Biol.Chem., 299, 2023
|
|
5O0P
 
 | |
6XPC
 
 | | Structure of human GGT1 in complex with full GSH molecule | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTATHIONE, ... | | Authors: | Terzyan, S.S, Hanigan, M. | | Deposit date: | 2020-07-08 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structures of glutathione- and inhibitor-bound human GGT1: critical interactions within the cysteinylglycine binding site.
J.Biol.Chem., 296, 2020
|
|
8BK5
 
 | | A structure of the truncated LpMIP with bound inhibitor JK095. | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-(hydroxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase, SODIUM ION | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
6XQP
 
 | | Structure of human D462-E4 TCR in complex with human MR1-5-OP-RU | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, BROMIDE ION, Beta-2-microglobulin, ... | | Authors: | Awad, W, Rossjohn, J. | | Deposit date: | 2020-07-10 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Atypical TRAV1-2 - T cell receptor recognition of the antigen-presenting molecule MR1.
J.Biol.Chem., 295, 2020
|
|
6QGD
 
 | | Structure of human Mcl-1 in complex with thienopyrimidine inhibitor | | Descriptor: | 2-[(6-ethyl-5-phenyl-thieno[2,3-d]pyrimidin-4-yl)amino]-3-oxidanyl-propanoic acid, Maltose-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
8BGY
 
 | | O-Methyltransferase Plu4890 in complex with SAH and AQ-284a | | Descriptor: | 1,3-dimethoxy-8-oxidanyl-anthracene-9,10-dione, CHLORIDE ION, Methyltransferase Plu4890, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2022-10-28 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A set of closely related methyltransferases for site-specific tailoring of anthraquinone pigments.
Structure, 31, 2023
|
|
8BIB
 
 | | O-Methyltransferase Plu4890 in complex with SAH and AQ-256 | | Descriptor: | 1,3,8-tris(oxidanyl)anthracene-9,10-dione, CARBONATE ION, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2022-11-02 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A set of closely related methyltransferases for site-specific tailoring of anthraquinone pigments.
Structure, 31, 2023
|
|
8BIJ
 
 | | O-Methyltransferase Plu4894 (mutant I88M, W91L, C97Y, S142L, G146V, Y258M, L270F, S309Y) in complex with SAH | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Methyltransferase Plu4894 mutant I88M, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2022-11-02 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A set of closely related methyltransferases for site-specific tailoring of anthraquinone pigments.
Structure, 31, 2023
|
|
8BIC
 
 | | O-Methyltransferase Plu4891 in complex with SAH | | Descriptor: | GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2022-11-02 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A set of closely related methyltransferases for site-specific tailoring of anthraquinone pigments.
Structure, 31, 2023
|
|
8BIR
 
 | | O-Methyltransferase Plu4895 in complex with SAH and AQ-256 | | Descriptor: | 1,3,8-tris(oxidanyl)anthracene-9,10-dione, CHLORIDE ION, IODIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2022-11-02 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A set of closely related methyltransferases for site-specific tailoring of anthraquinone pigments.
Structure, 31, 2023
|
|
8BBS
 
 | | Structure of AKR1C3 in complex with a bile acid fused tetrazole inhibitor | | Descriptor: | (4~{R})-4-[(1~{R},2~{S},5~{R},6~{R},13~{S},14~{S},17~{R},19~{R})-6,14-dimethyl-17-oxidanyl-7,8,9,10-tetrazapentacyclo[11.8.0.0^{2,6}.0^{7,11}.0^{14,19}]henicosa-8,10-dien-5-yl]pentanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Petri, E.T, Skerlova, J, Marinovic, M, Brynda, J, Kugler, M, Skoric, D, Bekic, S, Celic, A.S, Rezacova, P. | | Deposit date: | 2022-10-14 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray structure of human aldo-keto reductase 1C3 in complex with a bile acid fused tetrazole inhibitor: experimental validation, molecular docking and structural analysis.
Rsc Med Chem, 14, 2023
|
|
8BGZ
 
 | | O-Methyltransferase Plu4890 (mutant H229N) in complex with SAH and AQ-256 | | Descriptor: | 1,3,8-tris(oxidanyl)anthracene-9,10-dione, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2022-10-28 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A set of closely related methyltransferases for site-specific tailoring of anthraquinone pigments.
Structure, 31, 2023
|
|
8BGX
 
 | | O-Methyltransferase Plu4890 in complex with SAH and AQ-270a | | Descriptor: | 1-methoxy-3,8-bis(oxidanyl)anthracene-9,10-dione, CHLORIDE ION, GLYCEROL, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2022-10-28 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A set of closely related methyltransferases for site-specific tailoring of anthraquinone pigments.
Structure, 31, 2023
|
|
8BIF
 
 | | O-Methyltransferase Plu4892 in complex with SAH | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2022-11-02 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A set of closely related methyltransferases for site-specific tailoring of anthraquinone pigments.
Structure, 31, 2023
|
|
6ZJQ
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E517Q in complex with galactose | | Descriptor: | ACETATE ION, Beta-galactosidase, MALONATE ION, ... | | Authors: | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mapping the Transglycosylation Relevant Sites of Cold-Adapted beta-d-Galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 21, 2020
|
|
8BXU
 
 | | Crystal structure of Odorant Binding Protein 5 from Anopheles gambiae (AgamOBP5) with MPD (2-Methyl-2,4-pentanediol) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-ETHOXYETHANOL, Odorant binding protein, ... | | Authors: | Liggri, P.G.V, Tsitsanou, K.E, Zographos, S.E. | | Deposit date: | 2022-12-09 | | Release date: | 2023-03-22 | | Last modified: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The structure of AgamOBP5 in complex with the natural insect repellents Carvacrol and Thymol: Crystallographic, fluorescence and thermodynamic binding studies.
Int.J.Biol.Macromol., 237, 2023
|
|
8BJ1
 
 | | Crystal structure of Medicago truncatula histidinol-phosphate aminotransferase (HISN6) in the open state | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, SULFATE ION, ... | | Authors: | Rutkiewicz, M, Ruszkowski, M. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Insights into the substrate specificity, structure, and dynamics of plant histidinol-phosphate aminotransferase (HISN6).
Plant Physiol Biochem., 196, 2023
|
|
6YZ2
 
 | | Full length Open-form Sodium Channel NavMs F208L | | Descriptor: | DODECAETHYLENE GLYCOL, HEGA-10, Ion transport protein, ... | | Authors: | Sula, A, Sait, L.G, Hollingworth, D, Wallace, B.A. | | Deposit date: | 2020-05-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cannabidiol interactions with voltage-gated sodium channels.
Elife, 9, 2020
|
|
